Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline
Potabattula R, Zacchini F, Ptak GE, Dittrich M, Müller T, El Hajj N, Hahn T, Drummer C, Behr R, Lucas-Hahn A, Niemann H, Schorsch M, Haaf T., 01.07.2020
Abstract
In somatic cells/tissues, methylation of ribosomal DNA (rDNA) increases with age and age-related pathologies, which has a direct impact on the regulation of nucleolar activity and cellular metabolism. Here, we used bisulfite pyrosequencing and show that methylation of the rDNA transcription unit including upstream control element (UCE), core promoter, 18S rDNA, and 28S rDNA in human sperm also significantly increases with donor's age. This positive correlation between sperm rDNA methylation and biological age is evolutionarily conserved among mammals with widely different life spans such as humans, marmoset, bovine, and mouse. Similar to the tandemly repeated rDNA, methylation of human α-satellite and interspersed LINE1 repeats, marmoset α-satellite, bovine alpha- and testis satellite I, mouse minor and major satellite, and LINE1-T repeats increases in the aging male germline, probably related to their sperm histone packaging. Deep bisulfite sequencing of single rDNA molecules in human sperm revealed that methylation does not only depend on donor's age, but also depend on the region and sequence context (A vs. G alleles). Both average rDNA methylation of all analyzed DNA molecules and the number of fully (>50%) methylated alleles, which are thought to be epigenetically silenced, increase with donor's age. All analyzed CpGs in the sperm rDNA transcription unit show comparable age-related methylation changes. Unlike other epigenetic aging markers, the rDNA clock appears to operate in similar ways in germline and soma in different mammalian species. We propose that sperm rDNA methylation, directly or indirectly, influences nucleolar formation and developmental potential in the early embryo.
Potabattula R, Zacchini F, Ptak GE, Dittrich M, Müller T, El Hajj N, Hahn T, Drummer C, Behr R, Lucas-Hahn A, Niemann H, Schorsch M, Haaf T. Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline. Aging Cell. 2020 Aug;19(8):e13181. doi: 10.1111/acel.13181. Epub 2020 Jul 1. PMID: 32608562; PMCID: PMC7431825.
Publication: https://doi.org/10.1111/acel.13181
 Disclaimer
Disclaimer
The publication Increasing methylation of sperm rDNA and other repetitive elements in the aging male mammalian germline by Potabattula R, Zacchini F, Ptak GE, Dittrich M, Müller T, El Hajj N, Hahn T, Drummer C, Behr R, Lucas-Hahn A, Niemann H, Schorsch M, Haaf T. is published under an open access license: https://creativecommons.org/licenses/by/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Male aging effects on sperm repetitive DNA methylation
Methylome: Bisulfite Pyrosequencing
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm | Adult | A mature male germ cell that develops from a spermatid. | human | 295 |
Images

Figure 1: Methylation of the rDNA transcription unit in human sperm increases with donor's age.
Scatter plots show significant (****p < 0.0001) positive correlations between donor's age (x-axis in years) and mean methylation (y-axis in %) of the UCE (26 CpGs), core promoter (9 CpGs), 18S rDNA (8 CpGs), and 28S rDNA (10 CpGs). One hundred and eighty-six sperm samples of cohort 1 (a) and 109 samples of cohort 2 (b) were analyzed by bisulfite pyrosequencing. Pearson's partial correlations were used to adjust for confounding factors
Licensed under: https://creativecommons.org/licenses/by/4.0/

Table 1:
Pearson's partial correlations between donor's age and mean repeat methylation in human sperm cohorts 1 and 2
Licensed under: https://creativecommons.org/licenses/by/4.0/

Table S1:
Clinical parameters of the analyzed human sperm samples
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 2: Sequence- and age-dependent rDNA methylation and epimutation rates
Methylome: Deep Bisulfite Sequencing
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm | 26 - 26 years | A mature male germ cell that develops from a spermatid. | Human | 23 | |
| CL_0000019: sperm | 43 - 60 years | A mature male germ cell that develops from a spermatid. | Human | 23 |
Images

Figure 2:
Allele-specific methylation of rDNA region 1 (comprising 38 CpGs in the external transcribed spacer) and region 2 (25 CpGs in the upstream control element and core promoter) in sperm samples from young and old human donors. Twenty-three informative samples from each age class were analyzed by deep bisulfite sequencing. Alleles with an A variant are indicated by a blue and alleles with a G variant by a green diamond symbol. Methylation of both alleles is increased in older males. Region 2 also shows a higher methylation of the A, compared to the G allele
Licensed under: https://creativecommons.org/licenses/by/4.0/

Figure S3:
Box plots presenting the epimutation rates (ERs) of alleles carrying the A versus the G variant in rDNA regions 1 (representing the external transcribed spacer) and 2 (upstream control element and core promoter). The bottom and the top of the box represent the 25th and 75 th percentile, respectively. The median is represented by a horizontal line. Bars extend from the boxes to at most 1.5 times the height of the box. Outliers are indicated by an open circle and extreme outliers by a star symbol. The p values indicate significant differences of the ERs between 23 young versus 23 old donors and between rDNA region 1 versus 2, respectively. Mann-Whitney U tests were used for group comparisons.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 3: rDNA methylation in cord blood
Methylome: Bisulfite Pyrosequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0004053: umbilical cord blood | newborn conceived by IVF/ICSI | Blood from the umbilical cord of a newborn baby. This blood contains high concentrations of stem cells. [database_cross_reference: Dictionary_of_Cancer_Terms:http://www.cancer.gov/] | Human | 121 |
| BTO_0004053: umbilical cord blood | newborn, independent cohort | Blood from the umbilical cord of a newborn baby. This blood contains high concentrations of stem cells. [database_cross_reference: Dictionary_of_Cancer_Terms:http://www.cancer.gov/] | Human | 109 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| UBERON_0012168: umbilical cord blood | newborn conceived by IVF/ICSI | Blood that remains in the placenta and in the attached umbilical cord after childbirth[WP]. | Human | 121 | |
| UBERON_0012168: umbilical cord blood | newborn, independent cohort | Blood that remains in the placenta and in the attached umbilical cord after childbirth[WP]. | Human | 109 |
Images

Figure S4: Methylation of the rDNA upstream control element (UCE) and promoter in human fetal cord blood.
Each dot represents one of 121 cord blood samples. The y-axis indicates mean methylation (in %) of 26 CpG and 9 CpG sites in the UCE and promoter, respectively. The x-axis indicates the age of the father (in years) at conception. There is no significant correlation (Pearson's r = 0.01; p = 0.92 and r = 0.06; p = 0.53, respectively) between cord blood rDNA methylation and paternal age.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 4: rDNA methylation peripheral blood
Methylome: Bisulfite Pyrosequencing
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000553: peripheral blood | 1-70 years (female) | Blood circulating throughout the body. | Human | 94 |
| BTO_0000553: peripheral blood | 1-70 years (male) | Blood circulating throughout the body. | Human | 94 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000081: blood cell | 1-70 years (female) | A cell found predominately in the blood. | Human | 94 | |
| CL_0000081: blood cell | 1-70 years (male) | female, | Human | 94 |
Images

Figure 3: Methylation of the rDNA core promoter and α-satellite DNA in human peripheral blood, determined by bisulfite pyrosequencing.
rDNA (9 CpGs) methylation increases, whereas α-satellite DNA (4 CpGs) methylation decreases with donor's age. Scatter plots show significant (*p < 0.05 and **p < 0.01) correlations in 94 samples each from males (green dots) and females (red dots). Pearson's correlations were used for statistical analysis
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 5: Sperm rDNA clock
Methylome: Bisulfite Pyrosequencing
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm | training data set (cohort 1 and 2) | Human | 278 | ||
| CL_0000019: sperm | test data set (cohort 3) | Human | 154 |
Images

Figure 4: Building the rDNA methylation clock.
Scatter plots showing the chronological age (y-axis in years) versus rDNA methylation age (x-axis in years) in the training (N = 278) and testing cohort (N = 154). Model performance on training cohort: mean squared error (MSE) = 16.30, median absolute difference (MAD) = 2.78, Pearson's r = 0.72; and on test cohort: MSE = 16.96, MAD = 2.91, r = 0.67
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 6: Evolutionary conservation of age-related sperm rDNA methylation
Methylome: Bisulfite Pyrosequencing
Species
| Species |
|---|
| Mouse |
| Bovine |
| Marmoset |
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm | adult, 3- to 12-month-old mice | 80 samples of each month of 3- to 12-month-old mice | Mouse | 8 | |
| CL_0000019: sperm | adult, different ages | 36 sperm samples from 15 different bulls with different ages | Bovine | 15 | |
| CL_0000019: sperm | adult, 1- to 12-years | - | Marmoset | 16 | |
| CL_0000019: sperm | adult | cohort 1 and 2 | Human | 295 |
Images

Figure 5: Repeat methylation in mouse sperm increases with donor's age.
Scatter plots show significant (*p < 0.05, ***p < 0.001, and ****p < 0.0001) Pearson's correlations between donor's age (x-axis in years) and mean methylation (y-axis in %) of the rDNA spacer promoter (14 CpGs), rDNA gene promoter (7 CpGs), 18S rDNA (8 CpGs), 28S rDNA (10 CpGs), minor satellite DNA (2 CpGs), major satellite DNA (3 CpGs), and LINE1-T repeats (4 CpGs). Eighty sperm samples from 3- to 12-month-old mice were analyzed by bisulfite pyrosequencing
Licensed under: https://creativecommons.org/licenses/by/4.0/

Figure 6: Sperm methylation of 18S rDNA (8 CpGs), 28S rDNA (10 CpGs), bovine alpha-satellite (12 CpGs), and bovine testis satellite I (9 CpGs) increases with donor's age.
The color-coded regression lines show the age-related increase in DNA methylation of sperm samples from 12 bulls, analyzed by BPS at 2–3 different ages
Licensed under: https://creativecommons.org/licenses/by/4.0/
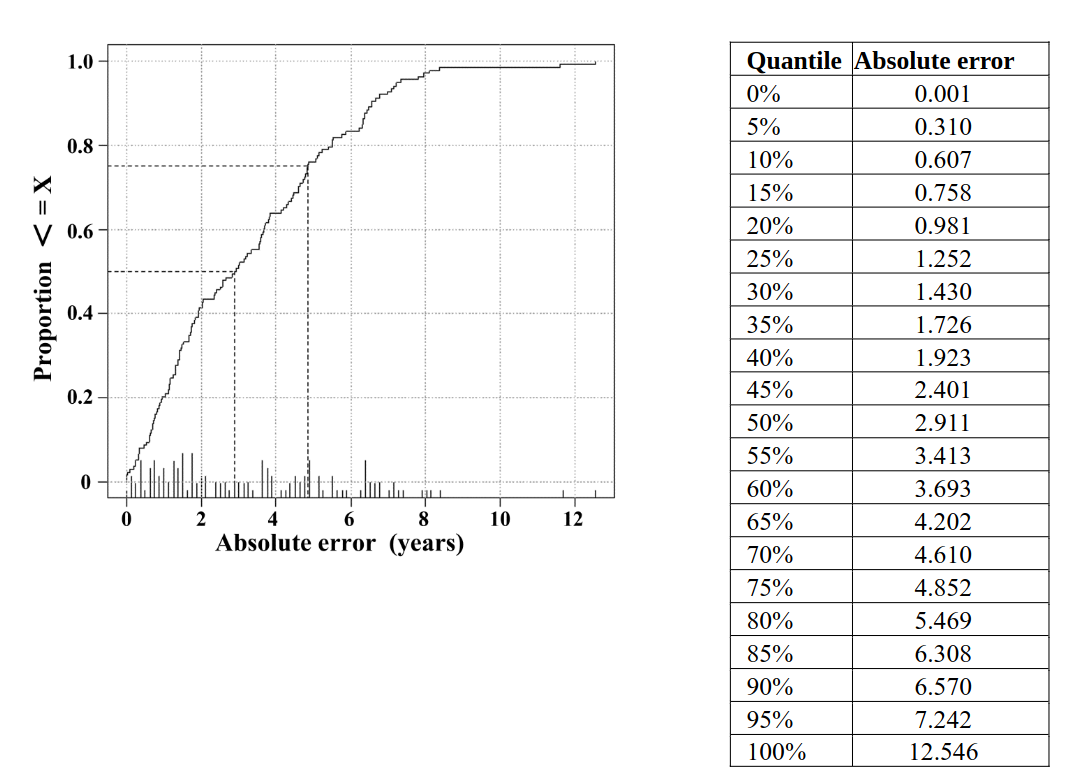
Figure S5: Distribution of absolute error in the test data set.
In total 154 test samples were analyzed. The cumulative distribution (i.e. the fraction of values < = x) is plotted on the y-axis versus the absolute error in years on the x-axis. Reference lines (dotted) are given for the 50% and 75% quantiles.
Licensed under: https://creativecommons.org/licenses/by/4.0/

Figure S7: Evolutionary conservation of the paternal age effect on sperm methylation of orthologous rDNA regions.
Scatter plots show positive correlations between donor's age (x-axis in percentage of lifespan) and mean methylation (y-axis in %) of 18S rDNA (8 CpGs) and 28S rDNA (10 CpGs), respectively, in 80 mouse (purple dots), 36 bull (yellow), 16 marmoset (red), and 295 human (green) sperm samples. Mean methylation of all 427 analyzed samples (indicated by the black regression lines) significantly increased with donor’s age (Pearson's r = 0.20; p < 0.0001 for 18S and r = 0.13; p < 0.01 for 28S rDNA)
Licensed under: https://creativecommons.org/licenses/by/4.0/
