Genome-wide DNA methylation changes in human spermatogenesis
Siebert-Kuss LM, Dietrich V, Di Persio S, Bhaskaran J, Stehling M, Cremers JF, Sandmann S, Varghese J, Kliesch S, Schlatt S, Vaquerizas JM, Neuhaus N, Laurentino S, 06.06.2024
Abstract
Sperm production and function require the correct establishment of DNA methylation patterns in the germline. Here, we examined the genome-wide DNA methylation changes during human spermatogenesis and its alterations in disturbed spermatogenesis. We found that spermatogenesis is associated with remodeling of the methylome, comprising a global decline in DNA methylation in primary spermatocytes followed by selective remethylation, resulting in a spermatids/sperm-specific methylome. Hypomethylated regions in spermatids/sperm were enriched in specific transcription factor binding sites for DMRT and SOX family members and spermatid-specific genes. Intriguingly, while SINEs displayed differential methylation throughout spermatogenesis, LINEs appeared to be protected from changes in DNA methylation. In disturbed spermatogenesis, germ cells exhibited considerable DNA methylation changes, which were significantly enriched at transposable elements and genes involved in spermatogenesis. We detected hypomethylation in SVA and L1HS in disturbed spermatogenesis, suggesting an association between the abnormal programming of these regions and failure of germ cells progressing beyond meiosis.
Siebert-Kuss LM, Dietrich V, Di Persio S, Bhaskaran J, Stehling M, Cremers JF, Sandmann S, Varghese J, Kliesch S, Schlatt S, Vaquerizas JM, Neuhaus N, Laurentino S. Genome-wide DNA methylation changes in human spermatogenesis. Am J Hum Genet. 2024 Jun 6;111(6):1125-1139. doi: 10.1016/j.ajhg.2024.04.017
Publication: https://doi.org/10.1016/j.ajhg.2024.04.017 Repository: https://github.com/VerenaDietrich/Spermatogenesis-Methylome
 Disclaimer
Disclaimer
The publication Genome-wide DNA methylation changes in human spermatogenesis by Siebert-Kuss LM, Dietrich V, Di Persio S, Bhaskaran J, Stehling M, Cremers JF, Sandmann S, Varghese J, Kliesch S, Schlatt S, Vaquerizas JM, Neuhaus N, Laurentino S is published under an open access license: https://creativecommons.org/licenses/by/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Whole-genome methylation sequencing of human germ cells from normal spermatogenesis
Methylome: EM-seq
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | adult | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 3 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000018: spermatid | A male germ cell that develops from the haploid secondary spermatocytes. Without further division, spermatids undergo structural changes and give rise to spermatozoa. | Human | |||
| CL_0000019: sperm | A mature male germ cell that develops from a spermatid. | Human |
Images
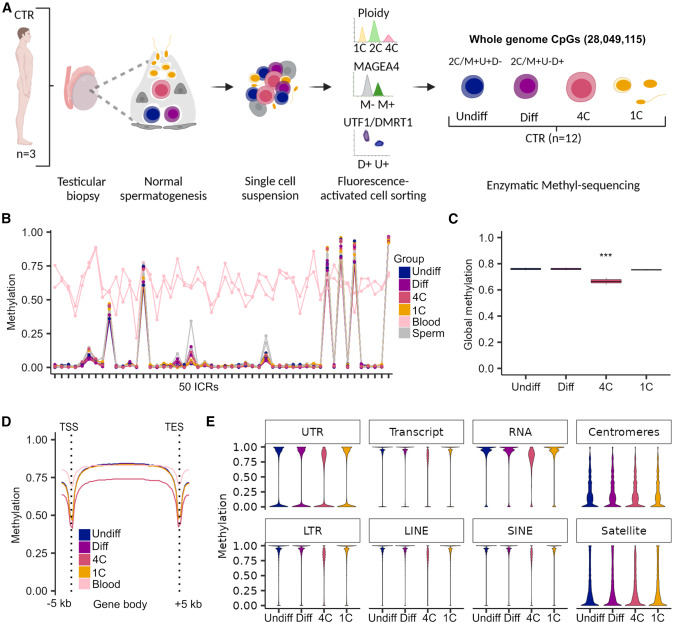
Figure 1. Primary spermatocytes exhibit genome-wide reduced DNA methylation levels
(A) Schematic illustration on the retrieval of whole-genome methylome data from germ cells of samples with normal spermatogenesis (control, CTR). CpGs refer to the mean CpG number captured by enzymatic methyl-sequencing in all germ cell fractions (n ¼ 12). (B) Line plot depicts the methylation in 50 imprinting control regions (ICRs) for each CTR sample and germ cell type compared to pub- lished blood and sperm samples.54. (C) Boxplots display the mean global DNA methylation levels. Statistical tests: ANOVA test followed by Tukey-HSD-test: *** <0.001 of 4C compared to all other germ cells. Data are represented as median (center line), upper/lower quartiles (box limits), 1.53 interquartile range (whiskers). (D) Line plot shows mean methylation levels per group across gene bodies divided into 50 intervals (bins) and 5 kb upstream and down- stream of the transcriptional start sites (TSSs) and transcriptional end sites (TESs). (E) Violin plots represent methylated CpGs across different genomic compartments. Undiff, undifferentiated spermatogonia; Diff, differ- entiating spermatogonia; 4C, primary spermatocytes; 1C, spermatids/sperm.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 2: Whole-genome methylation sequencing of human germ cells from cryptozoospermic individuals
Methylome: EM-seq
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | adult | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 3 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000020: spermatogonium | An euploid male germ cell of an early stage of spermatogenesis. | Human | |||
| CL_0000017: spermatocyte | A male germ cell that develops from spermatogonia. The euploid primary spermatocytes undergo meiosis and give rise to the haploid secondary spermatocytes which in turn give rise to spermatids. | Human |
Images
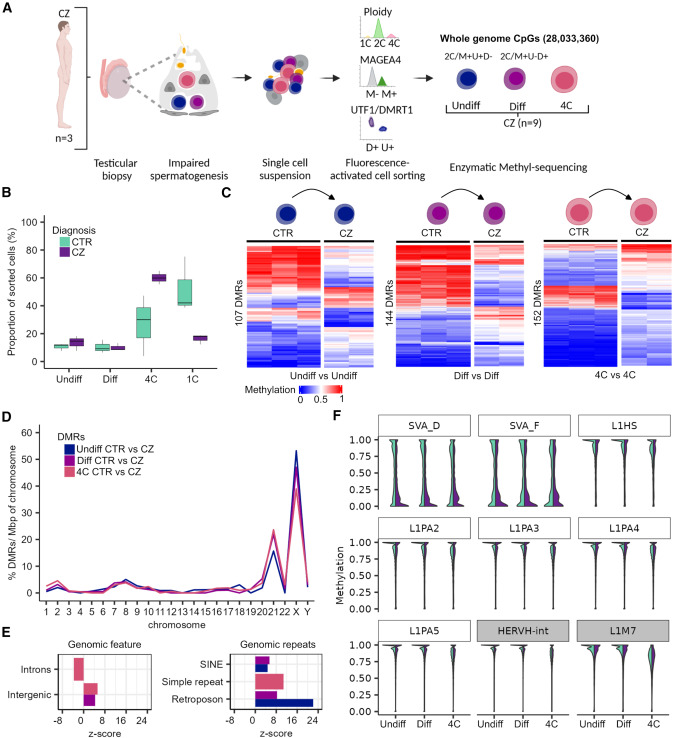
Figure 2. Disturbed spermatogenesis displays methylome changes at TEs and spermatogenesis genes
(A) Schematic illustration on the retrieval of whole-genome methylome data of germ cells from samples with disturbed spermatogenesis (CZ, cryptozoospermia). (B) Boxplots show the proportion of cell types among the sorted cells in the CTR and CZ subjects. Data are represented as median (center line), upper/lower quartiles (box limits), 1.53 interquartile range (whiskers). (C) Heatmaps display methylation values of the differentially methylated regions (DMRs) between CTR and CZ of the same cell type (Undiff vs. Undiff, Diff vs. Diff, and 4C vs. 4C). (D) Distribution of the CTR/CZ DMRs per chromosome scaled for chromosomal size (base pairs) and normalized by their total count within one group. (E) Enrichment of CTR/CZ DMRs for functional general genomic regions and genomic repeats. Positive and negative enrichments are indicated by Z score. Displayed annotations, p < 0.00019 by permutation tests. (F) Violin plots showing the CpG methylation of evolutionary younger (white boxes: L1Hs, L1PA2-5, and SVA_D/F) and older (gray boxes: HERVH-int and L1M7) TEs in CTR and CZ germ cells. Color coding of the group comparisons are depicted in (B). Undiff, undifferentiated spermatogonia; Diff, differentiating spermatogonia; 4C, primary spermatocytes; 1C, spermatids/sperm. (A) created with BioRender.com. See also Figures S4 and S5.
Licensed under: https://creativecommons.org/licenses/by/4.0/
