Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility
Stallmeyer B, Bühlmann C, Stakaitis R, Dicke A-K, Ghieh F, Meier L, Zoch A, MacLeod D M, Steingröver J, Okutman Ö, Fietz D, Pilatz A, Riera-Escamilla A, Xavier M J, Ruckert C, Di Persio S, Neuhaus N, Gurbuz A S, Şalvarci A, Le May N, McEleny K, Friedrich C, van der Heijden G, Wyrwoll M J, Kliesch S, Veltman J A, Krausz C, Viville S, Conrad D F, O’Carroll D, Tüttelmann F, 09.08.2024
Abstract
piRNAs are crucial for transposon silencing, germ cell maturation, and fertility in male mice. Here, we report on the genetic landscape of piRNA dysfunction in humans and present 39 infertile men carrying biallelic variants in 14 different piRNA pathway genes, including PIWIL1, GTSF1, GPAT2, MAEL, TDRD1, and DDX4. In some affected men, the testicular phenotypes differ from those of the respective knockout mice and range from complete germ cell loss to the production of a few morphologically abnormal sperm. A reduced number of pachytene piRNAs was detected in the testicular tissue of variant carriers, demonstrating impaired piRNA biogenesis. Furthermore, LINE1 expression in spermatogonia links impaired piRNA biogenesis to transposon de-silencing and serves to classify variants as functionally relevant. These results establish the disrupted piRNA pathway as a major cause of human spermatogenic failure and provide insights into transposon silencing in human male germ cells.
Stallmeyer, B., Bühlmann, C., Stakaitis, R. et al. Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility. Nat Commun 15, 6637 (2024). https://doi.org/10.1038/s41467-024-50930-9
Publication: https://doi.org/10.1038/s41467-024-50930-9
 Disclaimer
Disclaimer
The publication Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility by Stallmeyer B, Bühlmann C, Stakaitis R, Dicke A-K, Ghieh F, Meier L, Zoch A, MacLeod D M, Steingröver J, Okutman Ö, Fietz D, Pilatz A, Riera-Escamilla A, Xavier M J, Ruckert C, Di Persio S, Neuhaus N, Gurbuz A S, Şalvarci A, Le May N, McEleny K, Friedrich C, van der Heijden G, Wyrwoll M J, Kliesch S, Veltman J A, Krausz C, Viville S, Conrad D F, O’Carroll D, Tüttelmann F is published under an open access license:
https://creativecommons.org/licenses/by/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team
Relevant data sets presented in the publication have been identified. If possible, annotations (title, general
information, conditions, processed tissue types and processed cell types) have been added based on information from
the publication. Data tables and images that provide a good overview on the publication's findings on the data set
have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title
and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted
adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from
the publication.
Data set 1: Genetic variants in the piRNA pathway in the MERGE cohort
Exome: Exome Sequencing
Conditions
| Human phenotype ontology |
Participants |
Comment |
| HP:0000027: Azoospermia |
1448 |
Absence of any measurable level of sperm,whereby spermatozoa cannot be observed even after centrifugation of the semen pellet. |
| HP:0030974: Cryptozoospermia |
454 |
Sperm only identified after centrifugation of the ejaculate |
| HP:0034815: Extreme oligozoospermia |
158 |
Sperm count < 2 million |
| HP:0034818: Severe oligozoospermia |
67 |
Sperm count <10 million |
Tissue Types
| BRENDA tissue ontology |
Maturity |
Description |
Species |
Replicates |
| BTO_0000553: peripheral blood |
adult |
Blood circulating throughout the body. |
Human |
|
| BTO_0001363: testis |
average age: 34 |
A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. |
Human |
2352.0 |
Cell Types
| Cell ontology |
Maturity |
Description |
Species |
Replicates |
Cells per replicate |
| CL_0000020: spermatogonium |
|
An euploid male germ cell of an early stage of spermatogenesis. |
Human |
|
|
| CL_0000017: spermatocyte |
|
A male germ cell that develops from spermatogonia. The euploid primary spermatocytes undergo meiosis and give rise to the haploid secondary spermatocytes which in turn give rise to spermatids. |
Human |
|
|
| CL_0000018: spermatid |
|
A male germ cell that develops from the haploid secondary spermatocytes. Without further division, spermatids undergo structural changes and give rise to spermatozoa. |
Human |
|
|
Images
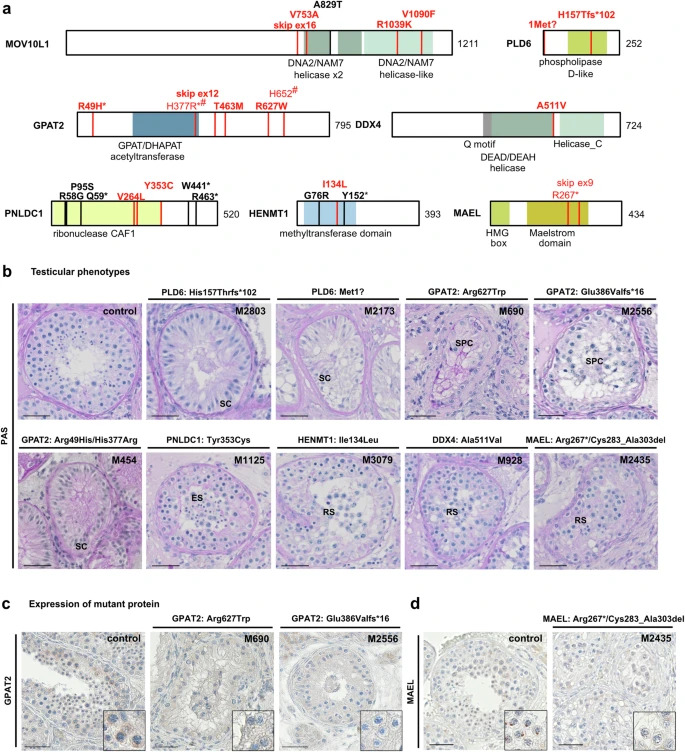
Figure 1: Biallelic variants identified in human piRNA biogenesis-associated genes
a Localization of variants in schematic of MOV10L1, PLD6, GPAT2, PNLDC1, MAEL, DDX4, and HENMT1 structure with protein domains colored and newly identified biallelic variants (red, bold for homozygous) as well as previously described homozygous variants (black) indicated. Pairs of compound heterozygous variants are indicated by identical symbols (*,#) in superscript. Helicase domains (green): DEAD/DEAH, Helicase_C, DNA2/NAM7; CAF1 chromatin assembly factor 1 domain (yellow); GPAT/DHAPAT acetyltransferase and methyltransferase domains (blue). b Periodic acid-Schiff (PAS) staining of representative testicular tissue of variant carriers demonstrating SCO in M2803 [PLD6, p.(His157Thrfs*102)], M2173 [PLD6, p.(Met1?)] and M454 [GPAT2, p.(His377Arg)/(Arg49His)] and presence of haploid germ cells (round/elongated spermatids) in M1125 [PNLDC1, p.(Tyr353Cys)], M3079 [HENMT1, p.(Ile134Leu)], M928 [DDX4, p.(Ala511Val)], and M2435 [MAEL, p.(Arg267*)/ p.(Cys283_Ala303del)]. Representative tubules showing the most advanced stage of spermatogenesis observed in three independent sections are shown. c Immunohistochemical (IHC) staining for GPAT2 in testicular tissue with full spermatogenesis (control) and GPAT2 variant carriers with meiotic arrest, [M690, p.(Arg627Trp), [M2556, p.(Glu386Valfs*16)]. In control tissue, GPAT2 is expressed in perinuclear structures in spermatocytes and this staining pattern is absent in M690 and M2556. Representative tubules showing the staining pattern observed in independent sections (control: N = 3, proband: N = 2) are shown. d IHC for MAEL in testicular tissue with full spermatogenesis (control) and M2435 with compound heterozygous presence of two MAEL LoF variants p.(Arg267*)/p.(Cys283_Ala303del). In control tissue, MAEL is expressed in perinuclear structures in spermatocytes and distinct condensed structures in round spermatids and this staining pattern is absent in the variant carrier. Representative tubules showing the staining pattern observed in independent sections (control: N = 3, proband: N = 2) are shown. Scale bar = 50 µm. SC Sertoli cell, SPC spermatocyte, RS round spermatid, ES elongated spermatid.
Source: Stallmeyer, B., Bühlmann, C., Stakaitis, R. et al. Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility. Nat Commun 15, 6637 (2024). https://doi.org/10.1038/s41467-024-50930-9
Licensed under: https://creativecommons.org/licenses/by/4.0/
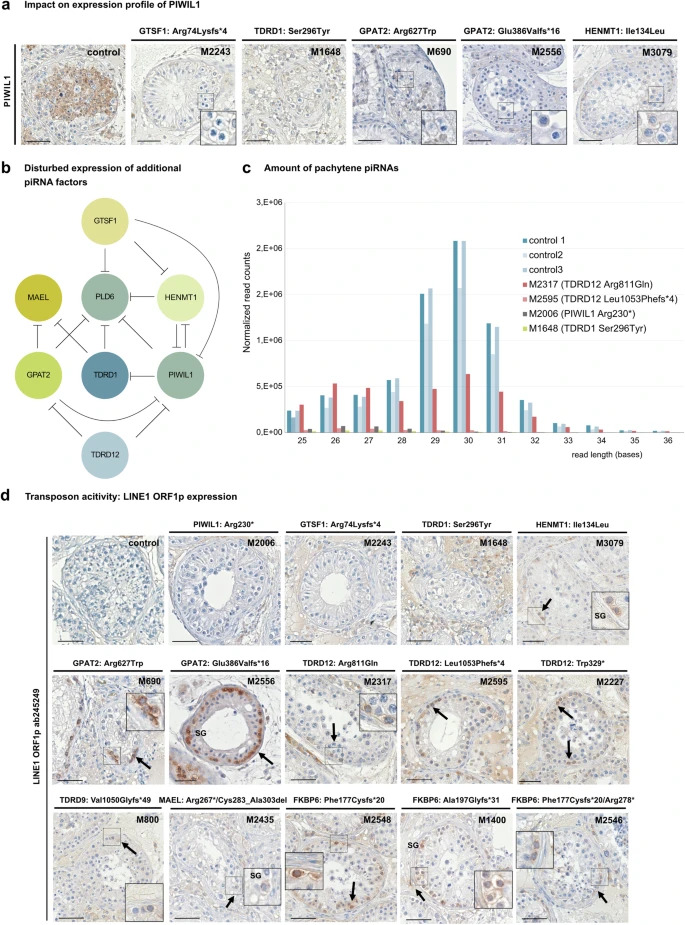
Figure 2: Functional impact of disturbed piRNA biogenesis
a Immunohistochemical staining demonstrating diminished expression of PIWIL1 in variant carriers M2243 [GTSF1, p.(Arg74Lysfs*4)], M1648 [TDRD1, p.(Ser296Tyr)], M690 [GPAT2, p.(Arg627Trp)], M2556, [GPAT2, p.(Glu386Valfs*16)] and M3079 [HENMT1, p.(Ile134Leu)]. Representative tubules showing the staining pattern observed in independent sections (control: N = 3, proband: N = 2) are shown. b Schematic depicting impact of loss of piRNA biogenesis protein function on expression of additional piRNA factors. Circles represent piRNA protein and inhibiting effects of loss-of-protein functions on the expression of further piRNA proteins are indicated. c Effect of biallelic variants in genes of the piRNA pathway on biogenesis of pachytene piRNAs. RNA isolated from snap frozen testicular tissue of M2006 [PIWIL1 p.(Arg230*)], M1648 [TDRD1 p.(Ser296Tyr)], M2317 [TDRD12 p.(Arg811Gln)] and M2595 [TDRD12 p.(Leu1053Phefs*4)] revealed a major loss of pachytene piRNAs with expected lengths of 26–31 bases when compared with controls with complete spermatogenesis (ctrl1-3; P = 0.000007). Shapiro-Wilk test was used to estimate the normality of the data. Since Shapiro-Wilk test indicated abnormal data distribution in both control and case groups, two-sided Mann-Whitney U test was used for comparing the expression changes of piRNAs with different length (26–31 nt) between both groups. Source data are provided as a Source Data file. d Immunohistochemical staining for LINE1 transposon specific protein LINE1 ORF1p in testicular tissue. LINE1 ORF1p was not detected in testicular tissue of controls with full spermatogenesis and PIWIL1, GTSF1, and TDRD1 variant carriers. In contrast, all three TDRD12 variant carriers, both GPAT2 variant carriers, and all three FKBP6 variant carriers revealed a concordant distinct and specific LINE1 ORF1p staining in spermatogonia. A similar effect was also seen in testicular tissue of MAEL, HENMT1, and TDRD9 variant carriers. Representative tubules showing the staining pattern observed in independent sections (control: N = 3, proband: N = 2) are shown. Scale bar = 50 µm. SG spermatogonia. b Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0
Source: Stallmeyer, B., Bühlmann, C., Stakaitis, R. et al. Inherited defects of piRNA biogenesis cause transposon de-repression, impaired spermatogenesis, and human male infertility. Nat Commun 15, 6637 (2024). https://doi.org/10.1038/s41467-024-50930-9
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 2: Genetic variants in the piRNA pathway in the MERGE cohort (Part 2)
Genome: Whole Genome Sequencing
Tissue Types
| BRENDA tissue ontology |
Maturity |
Description |
Species |
Replicates |
| BTO_0001363: testis |
average age: 34 |
A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. |
Human |
60 |
Data set 3: Genetic variants in the piRNA pathway in the Strasbourg cohort
Exome: Exome Sequencing
Conditions
| Human phenotype ontology |
Participants |
Comment |
| HP:0011961: Non-obstructive azoospermia |
23 |
Absence of any measurable level of sperm in his semen, resulting from a defect in the production of spermatozoa in the testes. This can be differentiated from obstructive azoospermia on the basis of testicular biopsy. |
Data set 4: Genetic variants in the piRNA pathway in the Barcelona cohort
Exome: Exome Sequencing
Conditions
| Human phenotype ontology |
Participants |
Comment |
| HP:0011961: Non-obstructive azoospermia |
235 |
Absence of any measurable level of sperm in his semen, resulting from a defect in the production of spermatozoa in the testes. This can be differentiated from obstructive azoospermia on the basis of testicular biopsy. |
Data set 5: Genetic variants in the piRNA pathway in the Nijmegen/Newcastle cohort
Exome: Exome Sequencing
Conditions
| Human phenotype ontology |
Participants |
Comment |
| HP:0000027: Azoospermia |
225 |
Absence of any measurable level of sperm,whereby spermatozoa cannot be observed even after centrifugation of the semen pellet. |
| HP:0030974: Cryptozoospermia |
41 |
A type of oligozoospermia in which spermatozoa can be detected in an ejaculate only after centrifugation and inspection of the pellet. |
 Disclaimer
Disclaimer

