Transcriptome analyses in infertile men reveal germ cell-specific expression and splicing patterns
Siebert-Kuss LM, Krenz H, Tekath T, Wöste M, Di Persio S, Terwort N, Wyrwoll MJ, Cremers JF, Wistuba J, Dugas M, Kliesch S, Schlatt S, Tüttelmann F, Gromoll J, Neuhaus N, Laurentino S, 29.11.2022
Abstract
The process of spermatogenesis-when germ cells differentiate into sperm-is tightly regulated, and misregulation in gene expression is likely to be involved in the physiopathology of male infertility. The testis is one of the most transcriptionally rich tissues; nevertheless, the specific gene expression changes occurring during spermatogenesis are not fully understood. To better understand gene expression during spermatogenesis, we generated germ cell-specific whole transcriptome profiles by systematically comparing testicular transcriptomes from tissues in which spermatogenesis is arrested at successive steps of germ cell differentiation. In these comparisons, we found thousands of differentially expressed genes between successive germ cell types of infertility patients. We demonstrate our analyses' potential to identify novel highly germ cell-specific markers (TSPY4 and LUZP4 for spermatogonia; HMGB4 for round spermatids) and identified putatively misregulated genes in male infertility (RWDD2A, CCDC183, CNNM1, SERF1B). Apart from these, we found thousands of genes showing germ cell-specific isoforms (including SOX15, SPATA4, SYCP3, MKI67). Our approach and dataset can help elucidate genetic and transcriptional causes for male infertility.
Lara M Siebert-Kuss, Henrike Krenz, Tobias Tekath, Marius Wöste, Sara Di Persio, Nicole Terwort, Margot J Wyrwoll, Jann-Frederik Cremers, Joachim Wistuba, Martin Dugas, Sabine Kliesch, Stefan Schlatt, Frank Tüttelmann, Jörg Gromoll, Nina Neuhaus, Sandra Laurentino Life Science Alliance Nov 2022, 6 (2) e202201633; DOI: 10.26508/lsa.202201633
Publication: http://doi.org/10.26508/lsa.202201633 Repository: https://ega-archive.org/studies/EGAS00001006135
 Disclaimer
Disclaimer
The publication Transcriptome analyses in infertile men reveal germ cell-specific expression and splicing patterns by Siebert-Kuss LM, Krenz H, Tekath T, Wöste M, Di Persio S, Terwort N, Wyrwoll MJ, Cremers JF, Wistuba J, Dugas M, Kliesch S, Schlatt S, Tüttelmann F, Gromoll J, Neuhaus N, Laurentino S is published under an open access license: https://creativecommons.org/licenses/by-nc-nd/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: RNA-sequencing of human testis with full spermatogenesis (CTR), spermatogonial arrest (SPG), spermatocyte arrest (SPC), round spermatid arrest (SPD), and Sertoli cell-only phenotype (SCO)
Transcriptome: RNA-Sequencing
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:0034299: Sertoli cell-only phenotype | 3 | Sertoli cells are found in the convoluted seminiferous tubules and form part of the blood-testis barrier in males. With Sertoli cell-only phenotype, spermatogenic cells, which are normally located in between the Seroli cells, are absent. |
| HP:4000187: Spermatogonial maturation arrest | 4 | A type of spermatogenesis maturation arrest in which the block of developmental occurs in the spermatogonial stage. Testicular histology shows seminiferous tubules with Sertoli cells and spermatogonial cells but no further differentiated cells like spermatocytes or round spermatids. |
| HP:0031039: Early spermatogenesis maturation arrest | 3 | A type of spermatogenesis maturation arrest in which the block of developmental occurs in the spermatocyte stage. Testicular histology shows seminiferous tubules with Sertoli cells, spermatogonial cells and spermatocytes but no further differentiated cells like round spermatids. |
| HP:0031040: Round spermatid arrest | 3 | A failure of spermatogenesis to progress beyond the round spermatid stage. The round spermatid is the first haploid cells produced during spermatogenesis and normally further develop into mature spermatozoa. This abnormality can be visualized with testicular biopsy and is characterized seminiferous tubules with increased numbers of round spermatids with few or no mature spermatozoa. |
| HP:control | 3 | Complete spermatogenesis as controls |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 16 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_4030036: early spermatid | round spermatids | Human | |||
| CL_4030037: late spermatid | elongated spermatids | Human | |||
| CL_0000017: spermatocyte | A male germ cell that develops from spermatogonia. The euploid primary spermatocytes undergo meiosis and give rise to the haploid secondary spermatocytes which in turn give rise to spermatids. | Human | |||
| CL_0000020: spermatogonium | An euploid male germ cell of an early stage of spermatogenesis. | Human |
Images
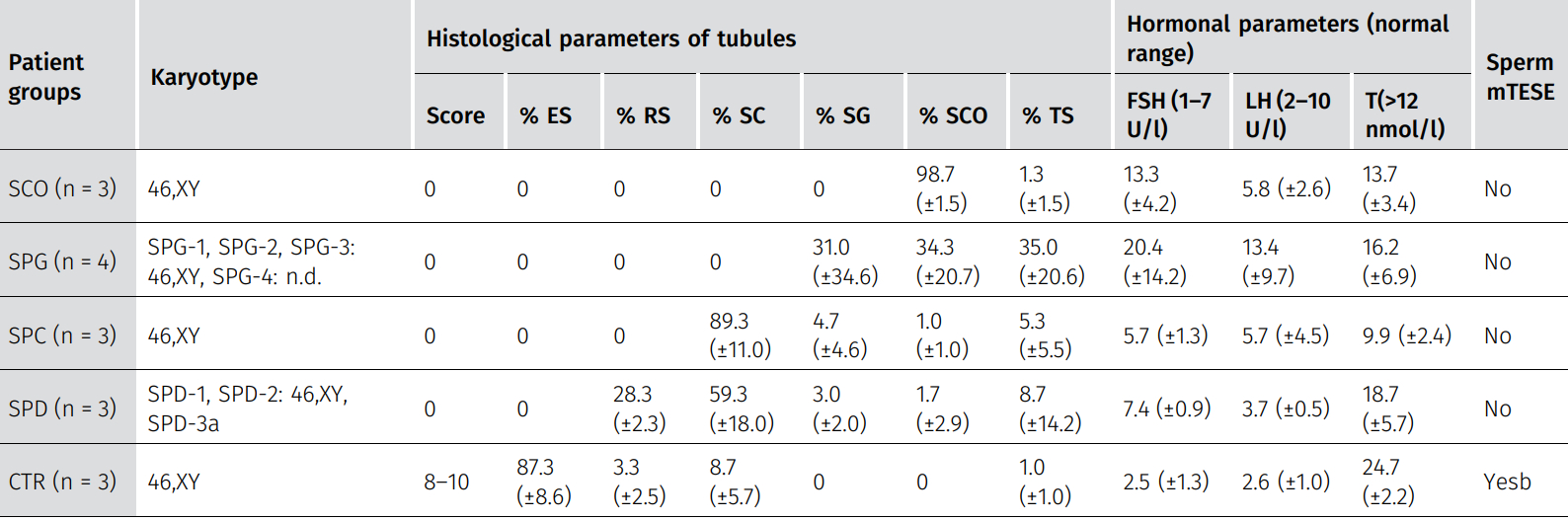
Table 1: Clinical characteristics of the patient groups.
Data are presented as mean ± SD. Percentage of tubules with the most advanced germ cell type present: elongated spermatids (%ES), round spermatids (%RS), spermatocytes (%SPC), spermatogonia (%SPG), Sertoli cell–only phenotype (%SCO), or tubular shadows (%TS). Score refers to Bergmann and Kliesch score (Bergmann & Kliesch, 2010). Hormonal parameters for follicle-stimulating hormone (FSH), luteinizing hormone (LH) and testosterone (T). a Patient SPD-3 had a low number of XXY karyotype mosaicism (47,XXY[2]/46,XY[28]). b Testicular sperm extraction (TESE) results: CTR-1 had 100/100 sperm, CTR-2 had an average of 89/100 sperm; no TESE result available for CTR-3 because the reason for surgery was a suspected malignant tumor. SCO, Sertoli cell–only; SPG, spermatogonial arrest; SPC, spermatocyte arrest; SPD, round spermatid arrest; CTR, control spermatogenesis; n.d., not determined.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
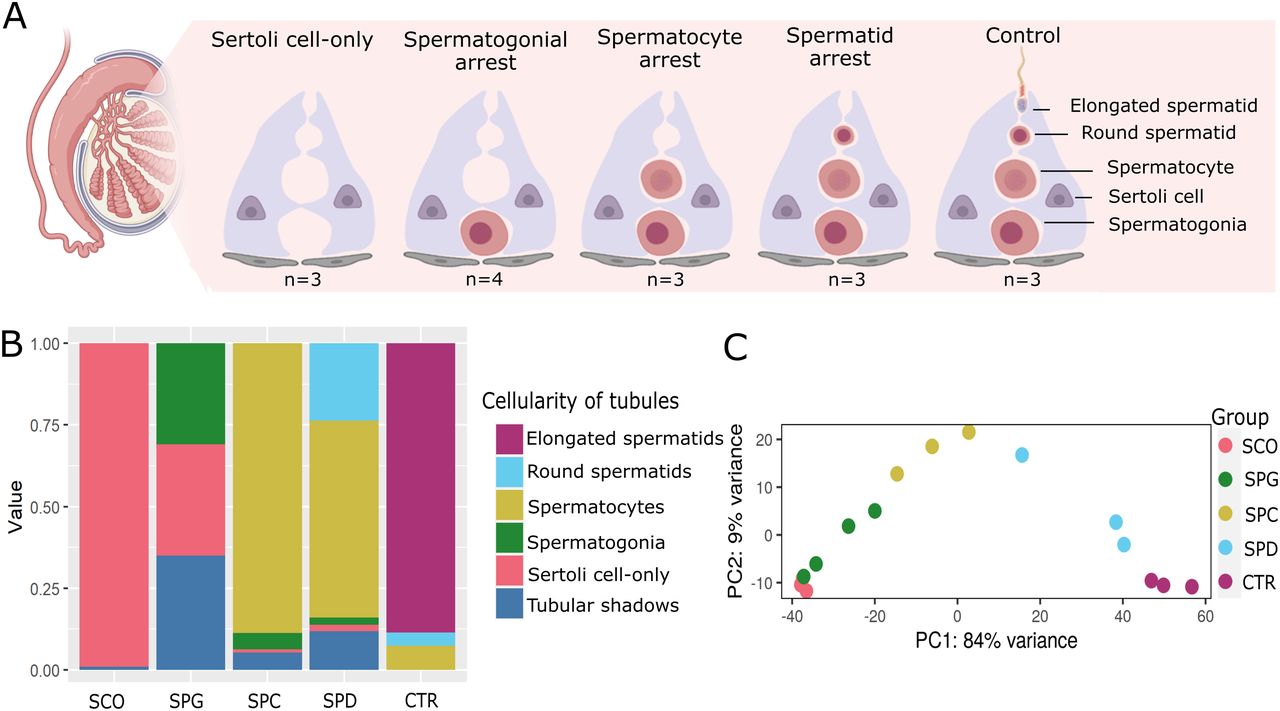
Figure 1: Cellular composition of the human testicular biopsies.
(A) Schematic illustration depicts the cellular composition of the testicular biopsies with Sertoli cell–only phenotype, arrest at the spermatogonial (SPG), spermatocyte (SPC), and spermatid (SPD) stage and samples with complete spermatogenesis, which were used as controls (CTR). (B) Stacked bar plots represent the proportional cellularity of round seminiferous tubules ranked according to the most advanced germ cell type in the tubule. The cellularity of samples from each group is averaged. (C) Principal component analysis (PCA) plot depicts clustering of the total RNA–sequenced samples based on the top 500 genes.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 2: Differentially expressed genes (DEGs) between SCO and SPG
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Images
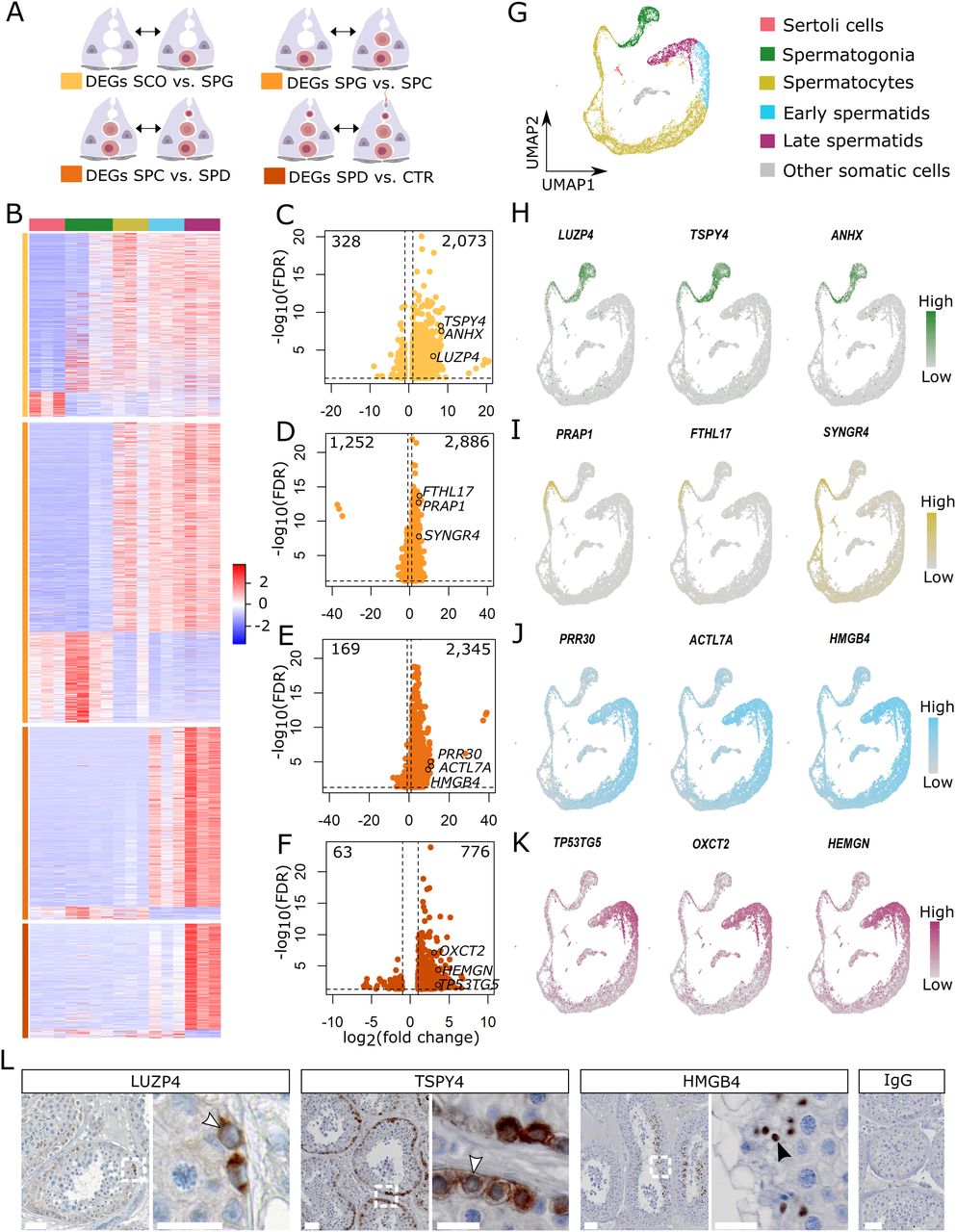
Figure 2: Examination of germ cell–specific gene expression.
(A) Schematic illustration of the group comparisons and the respective color codes of their differentially expressed genes (DEGs). (B) The heat map displays the normalized expression counts of the DEGs (rows) of each group comparisons across all samples (columns) scaled via a row Z-score. Red = increased; blue = decreased. (C, D, E, F) Volcano plots of the increased and decreased genes in samples with (C) spermatogonial, (D) spermatocyte, (E) and spermatid arrest and in (F) complete spermatogenesis. (G) UMAP plot depicts 15,546 cells integrated from three patients with obstructive azoospermia and complete spermatogenesis (Di Persio et al, 2021). (H, I, J, K) Feature plots show the expression of three novel genes for (H) spermatogonia, (I) spermatocytes, (J) round spermatids, and (K) elongated spermatids at single-cell level. (L) Micrographs showing immunohistochemical stainings for LUZP4, TSPY4, and HMGB4 in testicular tissue with full spermatogenesis (n = 3). Arrow heads in the inlays indicate positive spermatogonia (white) and round spermatids (black). IgG control shows no staining. Scale bars = 50 μm for micrographs and 20 μm for inlays. Data information: genes with a false discovery rate (FDR) < 0.05 and a log2 fold change (FC) ≥ 1 were considered DEGs based on Wald test and adjusted with Benjamini–Hochberg.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 3: DEGs between SPG and SPC
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Data set 4: DEGs between SPC and SPD
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Data set 5: DEGs between SPD and CTR
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Data set 6: Top 120 DEGs with absent or low expression in scRNA-seq datasets or different germ cell specificity
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Images
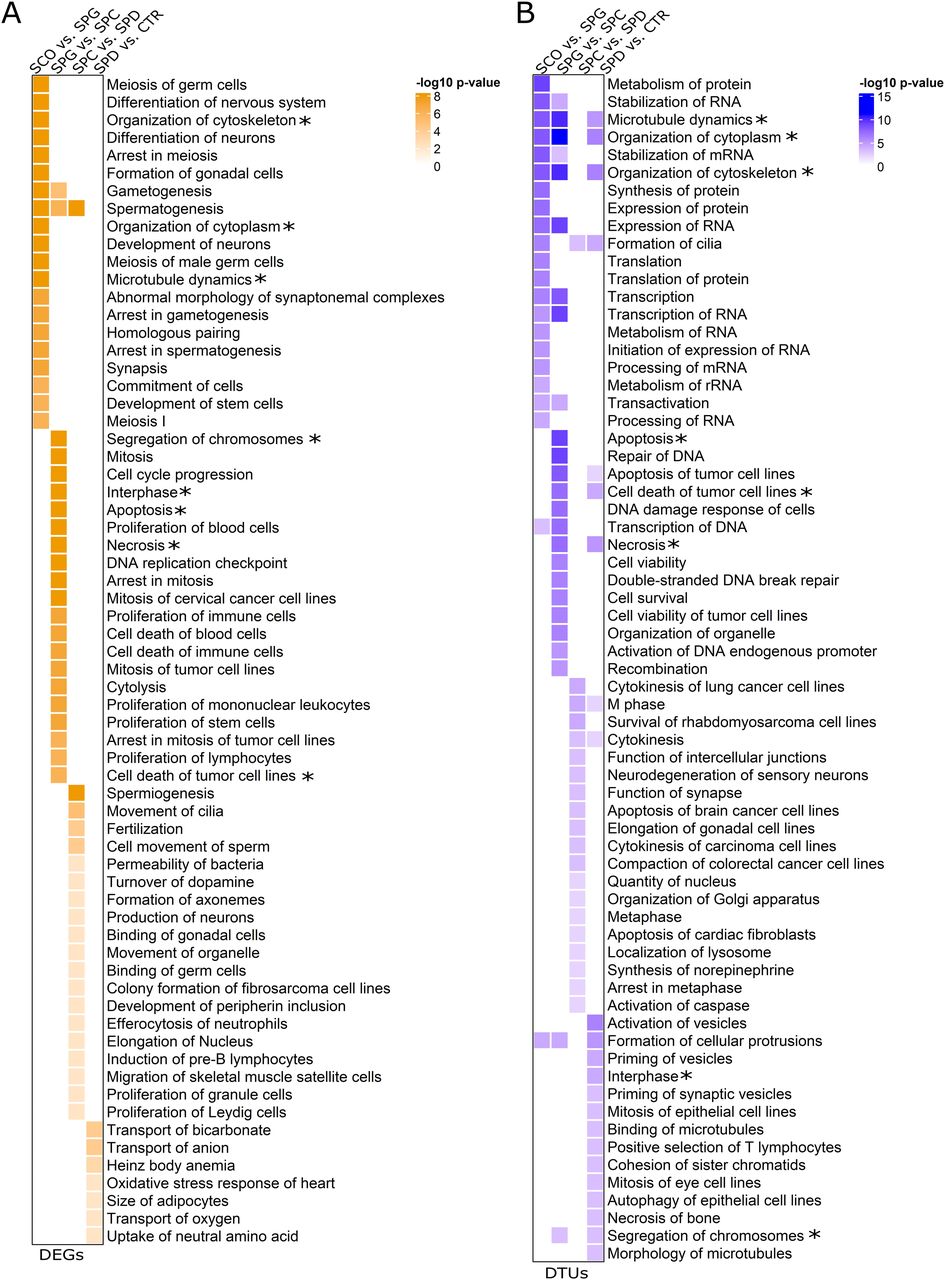
Figure 3: Molecular functions of differentially expressed genes (DEGs) and differential trascript usage (DTU) genes.
(A, B) Heat maps displaying the molecular functions revealed by IPA of all (A) DEGs and (B) DTU genes per group comparisons according to the −log 10 P-values. The top 20 molecular functions of each group comparison with P-values < 0.01 were included. * Molecular functions enriched in both the DEG and DTU gene sets
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 7: List of differential transcript usage (DTU) genes getween SCO and SPG
Transcriptome: Other
Species
| Species |
|---|
| Human |
Data set 8: List of DTU genes between SPG and SPC
Transcriptome: Other
Species
| Species |
|---|
| Human |
Data set 9: List of DTU genes between SPC and SPD
Transcriptome: Other
Species
| Species |
|---|
| Human |
Data set 10: List of DTU genes between SPD and CTR
Transcriptome: Other
Species
| Species |
|---|
| Human |
