TRIM71 deficiency causes germ cell loss during mouse embryogenesis and is associated with human male infertility
Lucia A. Torres-Fernández*, Jana Emich*, Yasmine Port*, Sibylle Mitschka, Marius Wöste, Simon Schneider, Daniela Fietz, Manon S. Oud, Sara di Persio, Nina Neuhaus, Sabine Kliesch, Michael Hölzel, Hubert Schorle, Corinna Friedrich, Frank Tüttelmann*, Waldemar Kolanus*. *These authors contributed equally., 13.05.2021
Abstract
Mutations affecting the germline can result in infertility or the generation of germ cell tumors (GCT), highlighting the need to identify and characterize the genes controlling germ cell development. The RNA-binding protein and E3 ubiquitin ligase TRIM71 is essential for embryogenesis, and its expression has been reported in GCT and adult mouse testes. To investigate the role of TRIM71 in mammalian germ cell embryonic development, we generated a germline-specific conditional Trim71 knockout mouse (cKO) using the early primordial germ cell (PGC) marker Nanos3 as a Cre-recombinase driver. cKO mice are infertile, with male mice displaying a Sertoli cell-only (SCO) phenotype which in humans is defined as a specific subtype of non-obstructive azoospermia characterized by the absence of germ cells in the seminiferous tubules. Infertility in male Trim71 cKO mice originates during embryogenesis, as the SCO phenotype was already apparent in neonatal mice. The in vitro differentiation of mouse embryonic stem cells (ESCs) into PGC-like cells (PGCLCs) revealed reduced numbers of PGCLCs in Trim71-deficient cells. Furthermore, TCam-2 cells, a human GCT-derived seminoma cell line which was used as an in vitro model for PGCs, showed proliferation defects upon TRIM71 knockdown. Additionally, in vitro growth competition assays, as well as proliferation assays with wild type and CRISPR/Cas9-generated TRIM71 mutant NCCIT cells showed that TRIM71 also promotes proliferation in this malignant GCTderived non-seminoma cell line. Importantly, the PGC-specific markers BLIMP1 and NANOS3 were consistently downregulated in Trim71 KO PGCLCs, TRIM71 knockdown TCam-2 cells and TRIM71 mutant NCCIT cells. These data collectively support a role for TRIM71 in PGC development. Last, via exome sequencing analysis, we identified several TRIM71 variants in a cohort of infertile men, including a loss-of-function variant in a patient with an SCO phenotype. Altogether, our work reveals for the first time an association of TRIM71 deficiency with human male infertility, and uncovers further developmental roles for TRIM71 in the germline during mouse embryogenesis.
TORRES-FERNÁNDEZ, Lucia A., et al. TRIM71 deficiency causes germ cell loss during mouse embryogenesis and is associated with human male infertility. Frontiers in cell and developmental biology, 2021, 9. Jg., S. 970.
Publication: https://doi.org/10.3389/fcell.2021.658966 Repository: https://www.biorxiv.org/content/10.1101/2021.02.01.429172v1
 Disclaimer
Disclaimer
The publication TRIM71 deficiency causes germ cell loss during mouse embryogenesis and is associated with human male infertility by Lucia A. Torres-Fernández*, Jana Emich*, Yasmine Port*, Sibylle Mitschka, Marius Wöste, Simon Schneider, Daniela Fietz, Manon S. Oud, Sara di Persio, Nina Neuhaus, Sabine Kliesch, Michael Hölzel, Hubert Schorle, Corinna Friedrich, Frank Tüttelmann*, Waldemar Kolanus*. *These authors contributed equally. is published under an open access license: https://creativecommons.org/licenses/by/4.0/. The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Figure 1. Trim71 is expressed in spermatogonial stem cells (SSCs) of adult fertile mice.
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| mouse |
Images
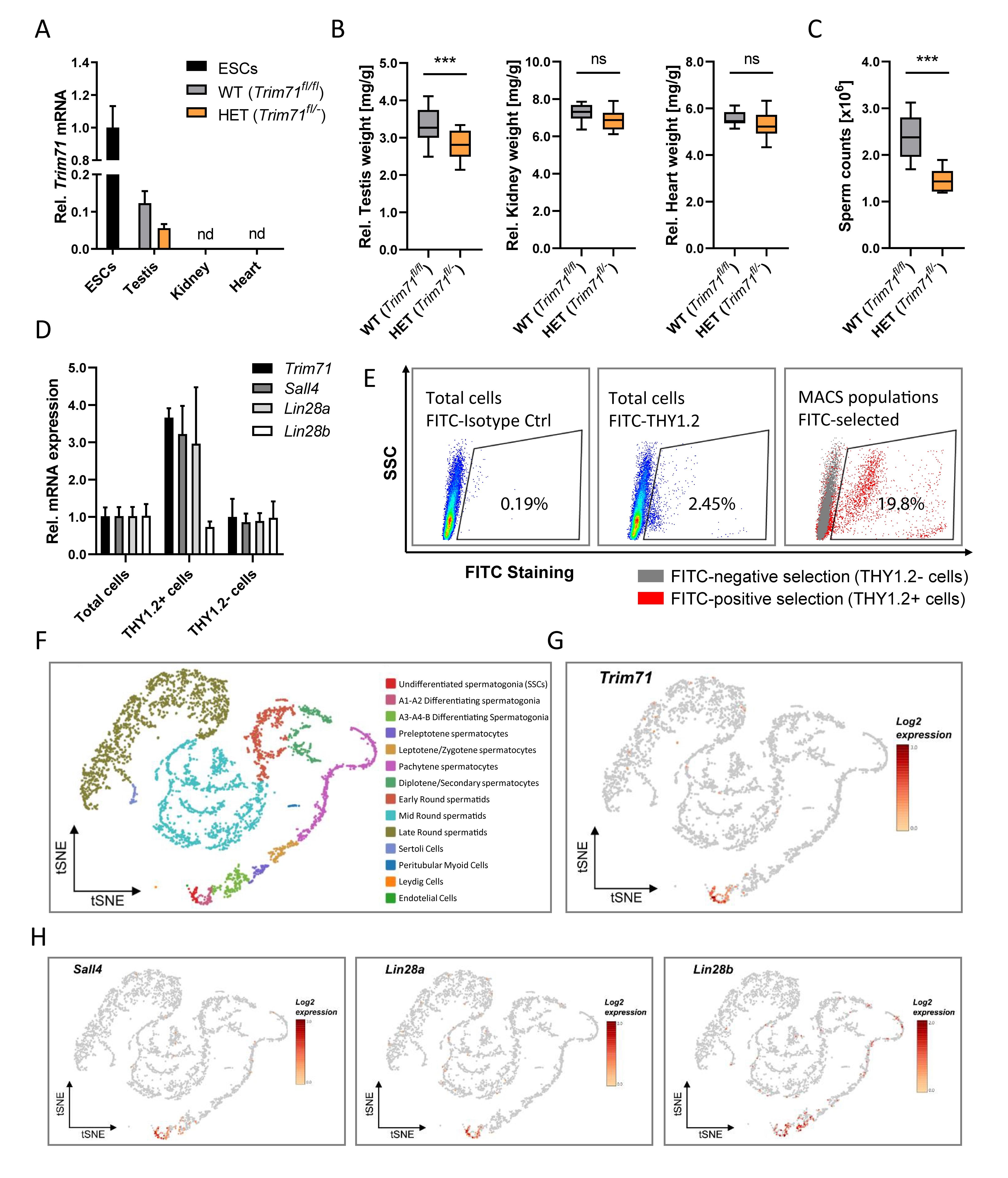
Figure 1. Trim71 is expressed in spermatogonial stem cells (SSCs) of adult fertile mice.
(A) qRT-PCR of Trim71 relative to Hprt housekeeping gene in testis, kidney and heart of wild type (WT, Trim71fl/fl ) and Trim71 heterozygous (HET, Trim71fl/-) adult male mice (10–14 weeks old), normalized to Trim71 relative levels in wild type murine ESCs (nd, not detected). Error bars represent SD (n = 3). (B) Organ weight [mg] relative to total body weight [g] of testis, kidney and heart of wild type (WT, Trim71fl/fl ) and Trim71 heterozygous (HET, Trim71fl/-) male adult mice (10–14 weeks old). Graphs represent Tukey plots (n = 14-16). ***P-value < 0.005, ns, non-significant (unpaired Student’s t-test). (C) Epididymal sperm counts in wild type (WT, Trim71fl/fl ) and Trim71 heterozygous (HET, Trim71fl/-) male adult mice (10–14 weeks old). ***P-value < 0.005 (unpaired Student’s t-test). (D) qRT-PCR of Trim71, the SSC markers Sall4 and Lin28a, and the differentiating spermatogonia marker Lin28b, relative to Hprt housekeeping gene in testes cell suspension before (total cells) and after THY1.2 MACS (THY1.2+/- cells). Error bars represent SD (n = 3). (E) Representative flow cytometry scatter plots of testes cell suspension before (total cells) and after THY1.2 MACS (THY1.2+/- cells). (F) tSNE plot (t-distributed stochastic neighbor embedding) of single-cell transcriptome data (scRNA-seq) from mouse testes as published by Hermann et al. (2018). Each dot represents a single cell and is colored according to its cluster identity as indicated on the figure key. (G) Expression pattern of Trim71 and (H) Sall4, Lin28a and Lin28b projected on the tSNE plot of the mouse scRNA-seq dataset. Red indicates high expression and gray indicates low or no expression, as shown by the figure key. See also Supplementary Figures 1, 2.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 2: Figure 2. Germline-specific Trim71 cKO male mice present a Sertoli cell-only (SCO) phenotype.
Other: Other
Species
| Species |
|---|
| mouse |
Images
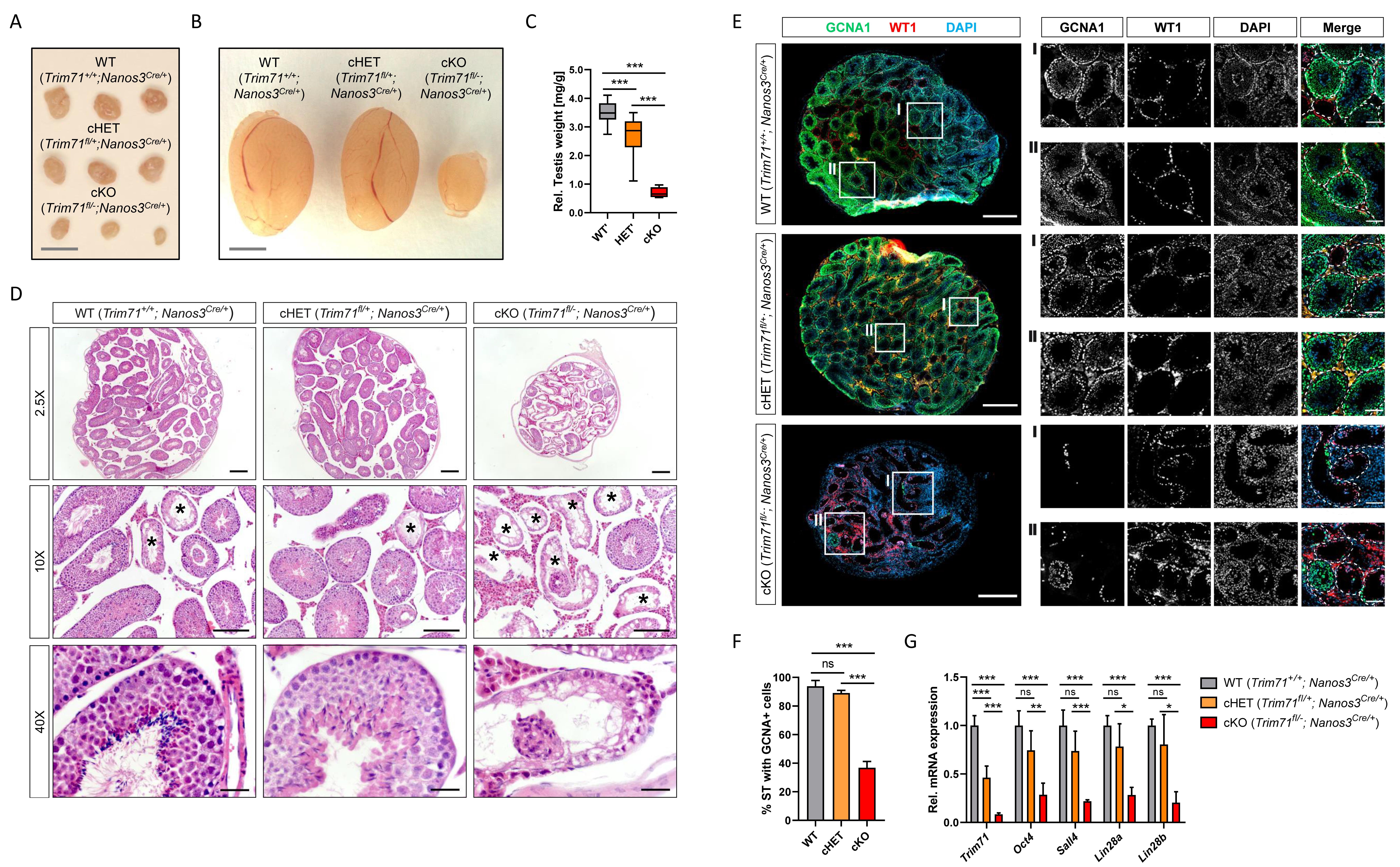
Figure 2. Germline-specific Trim71 cKO male mice present a Sertoli cell-only (SCO) phenotype.
(A) Representative images of ovaries and (B) testes of adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) mice. Scale bars represent 2 mm. (C) Testis weight [mg] relative to total body weight [g] of wild type (WT’), Trim71 heterozygous (HET’) and germline-specific Trim71 knockout (cKO) mice, summarized from Supplementary Figure 2C, joining several Nanos3 genotypes per Trim71 genotype. Error bars represent SD (n=8–29). ***P-value < 0.005 (one-way ANOVA, Tukey’s test). (D) Representative H&E stainings on paraffin testes cross-sections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) mice. Seminiferous tubules with a defective morphology are marked with an asterisk (*) in 10x magnification images. Scale bars represent 200, 100, and 20 µm in 2.5x, 10x, and 40x magnifications, respectively. (E) Representative immunofluorescence stainings on testes cryosections from adult wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) mice. Images show co-staining with GCNA1 (germ cells), WT1 (Sertoli cells) and DAPI (nuclei). For each genotype two regions – indicated as I and II – are depicted in higher magnification. Scale bars represent 500 µm for complete testes cross-sections and 100 µm for the magnification images. (F) Quantification of seminiferous tubules (ST) containing GCNA+ cells per testis cross-section from E, depicted as percentages. Error bars represent SD (n = 3). ***P-value < 0.005, ns, non-significant (unpaired Student’s t-test). (G) qRT-PCR of Trim71, the murine SSC markers Sall4, Lin28a and Oct4, and the differentiating spermatogonia marker Lin28b, relative to Hprt housekeeping gene in whole testis RNA of wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) male adult mice. Error bars represent SD (n=3-6). ***P-value < 0.005; ***P-value < 0.01; *P-value < 0.05; ns, non-significant (unpaired Student’s t-test). See also Supplementary Figures 3, 4.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 3: Figure 3. Infertility in germline-specific Trim71 cKO male mice has an embryonic origin.
Proteome: Other
Species
| Species |
|---|
| mouse |
Images
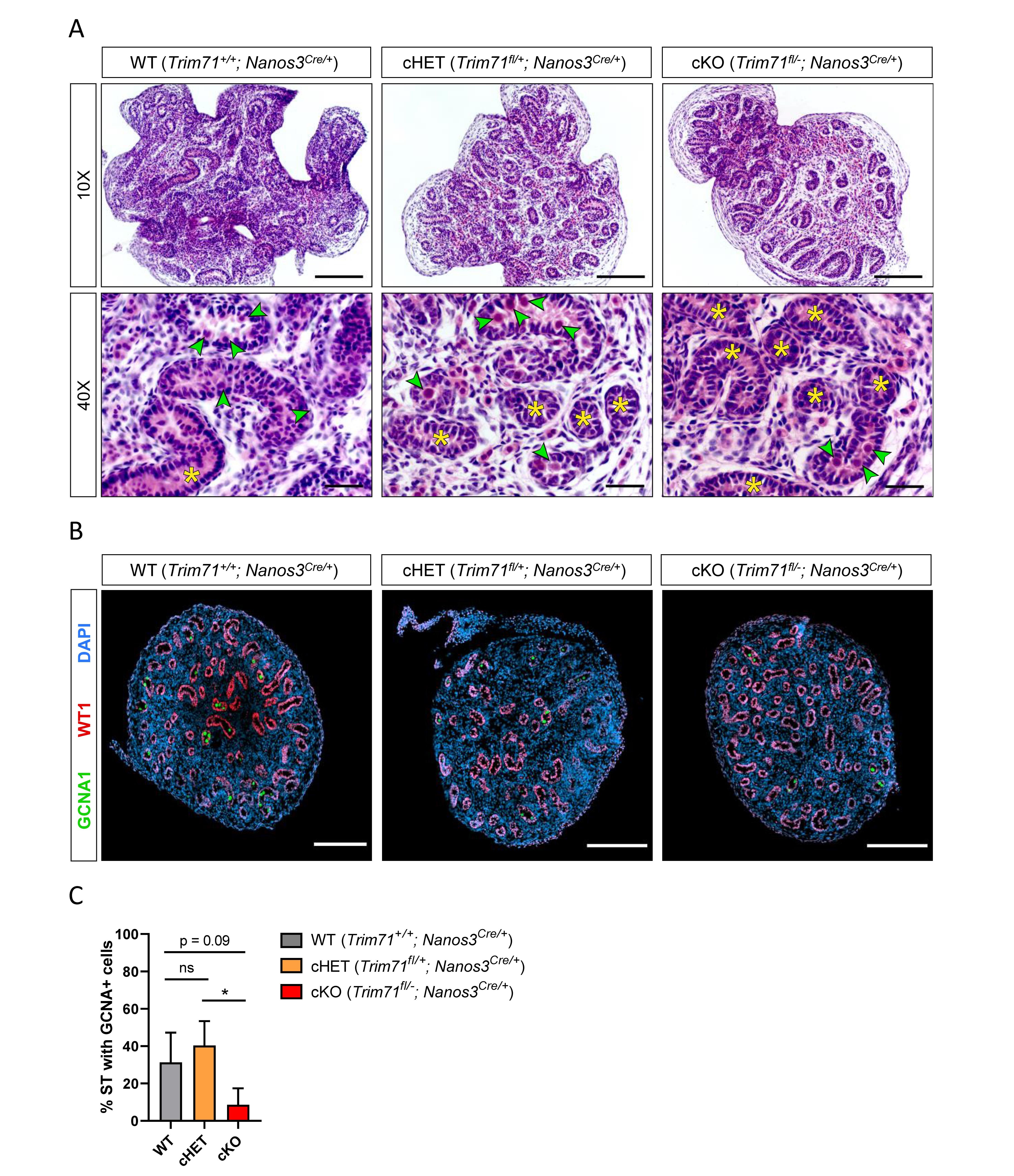
Figure 3. Infertility in germline-specific Trim71 cKO male mice has an embryonic origin.
(A) Representative H&E stainings on paraffin testes cross-sections from neonatal (P0.5) wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) male mice. Gonocytes within the seminiferous tubules are marked with a green arrow head, and gonocyte-lacking seminiferous tubules are marked with a yellow asterisk (*) in 40x magnification images. Scale bars represent 100 and 20 µm in 10x and 40x magnifications, respectively. (B) Representative immunofluorescence stainings on testes cryo-sections from neonatal (P0.5) wild type (WT, Trim71+/+; Nanos3Cre/+), germline-specific Trim71 heterozygous (cHET, Trim71fl/+; Nanos3Cre/+) and germline-specific Trim71 knockout (cKO, Trim71fl/-; Nanos3Cre/+) male mice. Images show co-staining with GCNA1 (gonocytes), WT1 (Sertoli cells) and DAPI (nuclei). Scale bars represent 200 µm. (C) Quantification of seminiferous tubules (ST) containing GCNA+ cells per testis cross-section from (B), depicted as percentages. Error bars represent SD (n=3). *P-value < 0.05, ns, non-significant (unpaired Student’s t-test). See also Supplementary Figures 4–6.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 4: Figure 4. TRIM71 controls proliferation in the in vitro PGC-like model cell line TCam-2.
Transcriptome: Other
Species
| Species |
|---|
| human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| human |
Images
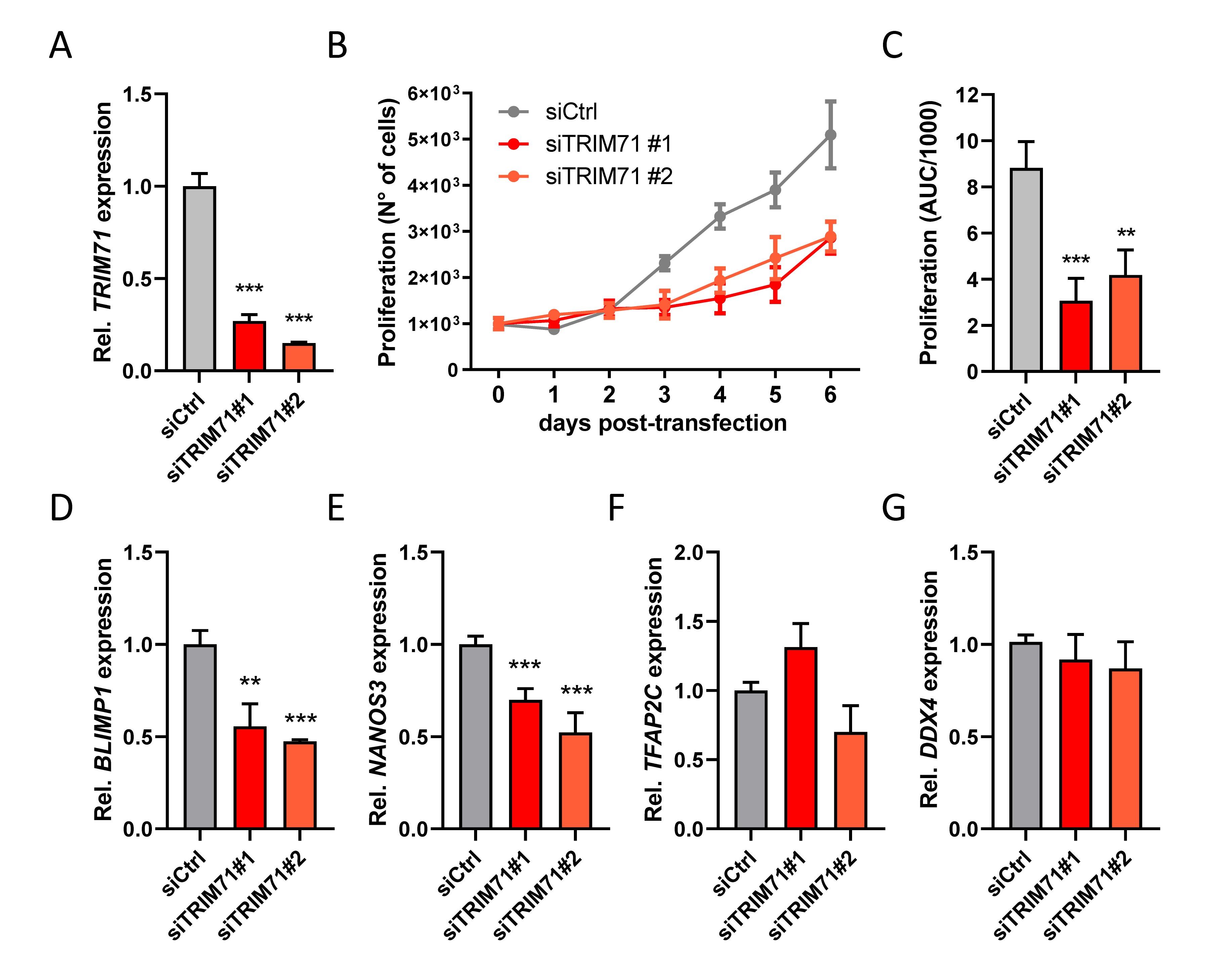
Figure 4. TRIM71 controls proliferation in the in vitro PGC-like model cell line TCam-2.
(A) RT-qPCR measurement of TRIM71 in wild type (siCtrl) and TRIM71 KD (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells 72 h post-transfection (n = 3). (B) Proliferation of wild type (siCtrl) and TRIM71 KD (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells depicting the increase in cell numbers over time after transfecting them with siRNAs and seeding them in equal numbers (d0). (n = 5). (C) Area under curve (AUC) measurements for the proliferation curves depicted in (B) (n = 5). (D–G) RT-qPCR measurements for the indicated PGC markers in wild type (siCtrl) and TRIM71 knockdown (siTRIM71 #1 and siTRIM71 #2) TCam-2 cells 72 h post-transfection (n=3–6). For all RT-qPCRs, the housekeeping gene 18S rRNA was used for normalization. For all graphs, error bars represent the SEM. ***P-value < 0.005, **P-value < 0.01 (unpaired Student’s t-test). See also Supplementary Figure 8.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 5: Figure 5. NCCIT cells with TRIM71 mutations show population maintenance defects in growth competition assays.
Genome: Targeted Genotyping
Species
| Species |
|---|
| human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| human |
Images
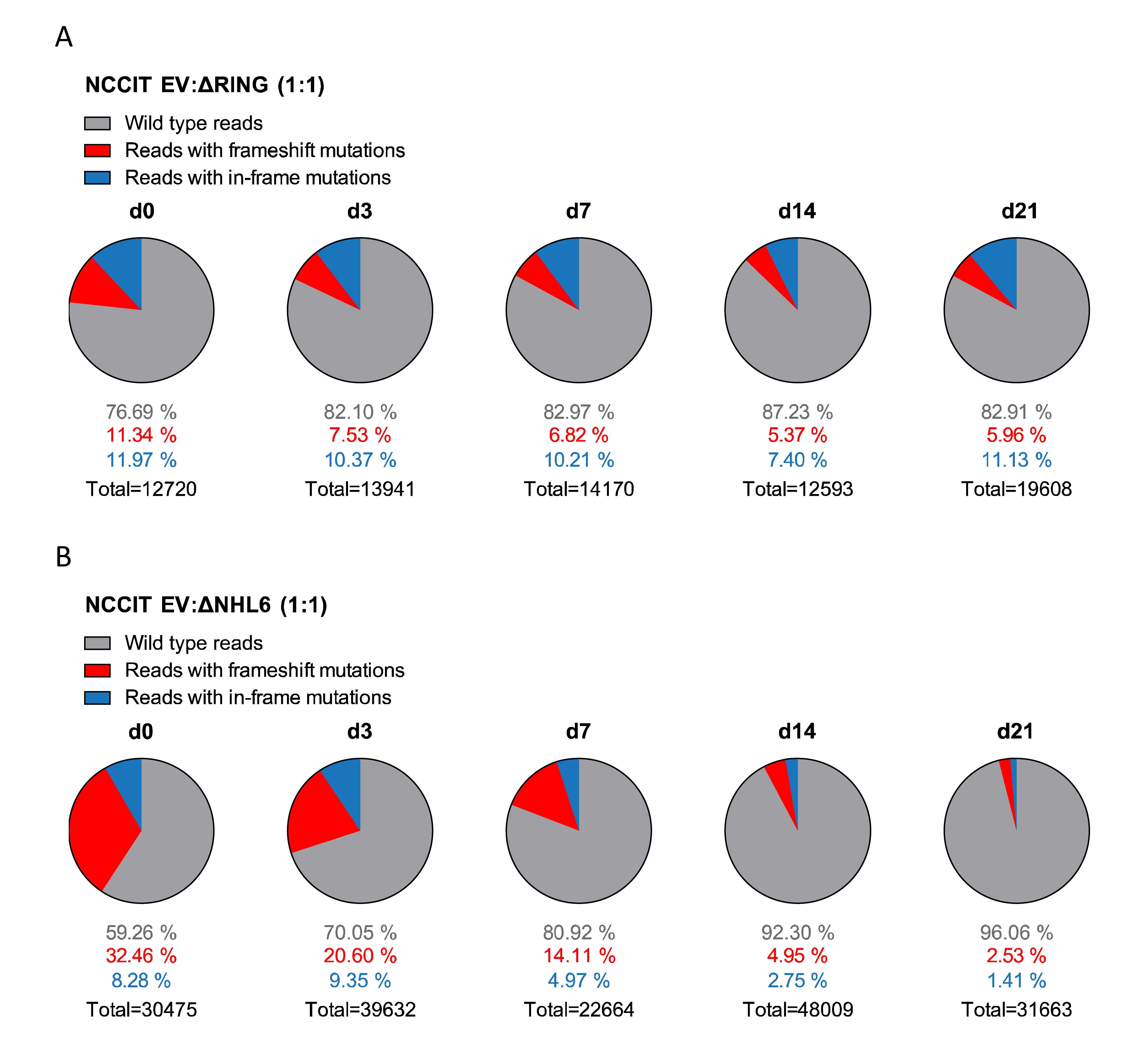
Figure 5. NCCIT cells with TRIM71 mutations show population maintenance defects in growth competition assays.
(A) Pie charts showing allele frequencies for wild type (EV) and TRIM71 RING mutant (deltaRING) or (B) TRIM71 NHL mutant (deltaNHL6) NCCIT cells obtained at different time points during growth competition assays. For the assay, NCCIT wild type cells (EV) were mixed 1:1 with either TRIM71 RING mutant (deltaRING) or TRIM71 NHL mutant (deltaNHL6) NCCIT cells and directly analyzed (d0) before their culturing, followed by subsequent analysis at several time points (d3, d7, d14, and d21). Allele frequency was analyzed via NGS using the Illumina MiSeq platform. For TRIM71 mutant alleles, reads with in-frame mutations and frameshift mutations (loss-of-function) in each respective domain (RING or NHL) are depicted. The total number of sequencing reads for each time point is indicated under each respective pie chart. See also Supplementary Figures 8–10.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 6: Figure 6. Exome sequencing in infertile SCO patients reveals an association of TRIM71 deficiency with human male infertility.
Exome: Whole Exome Sequencing
Species
| Species |
|---|
| human |
Images
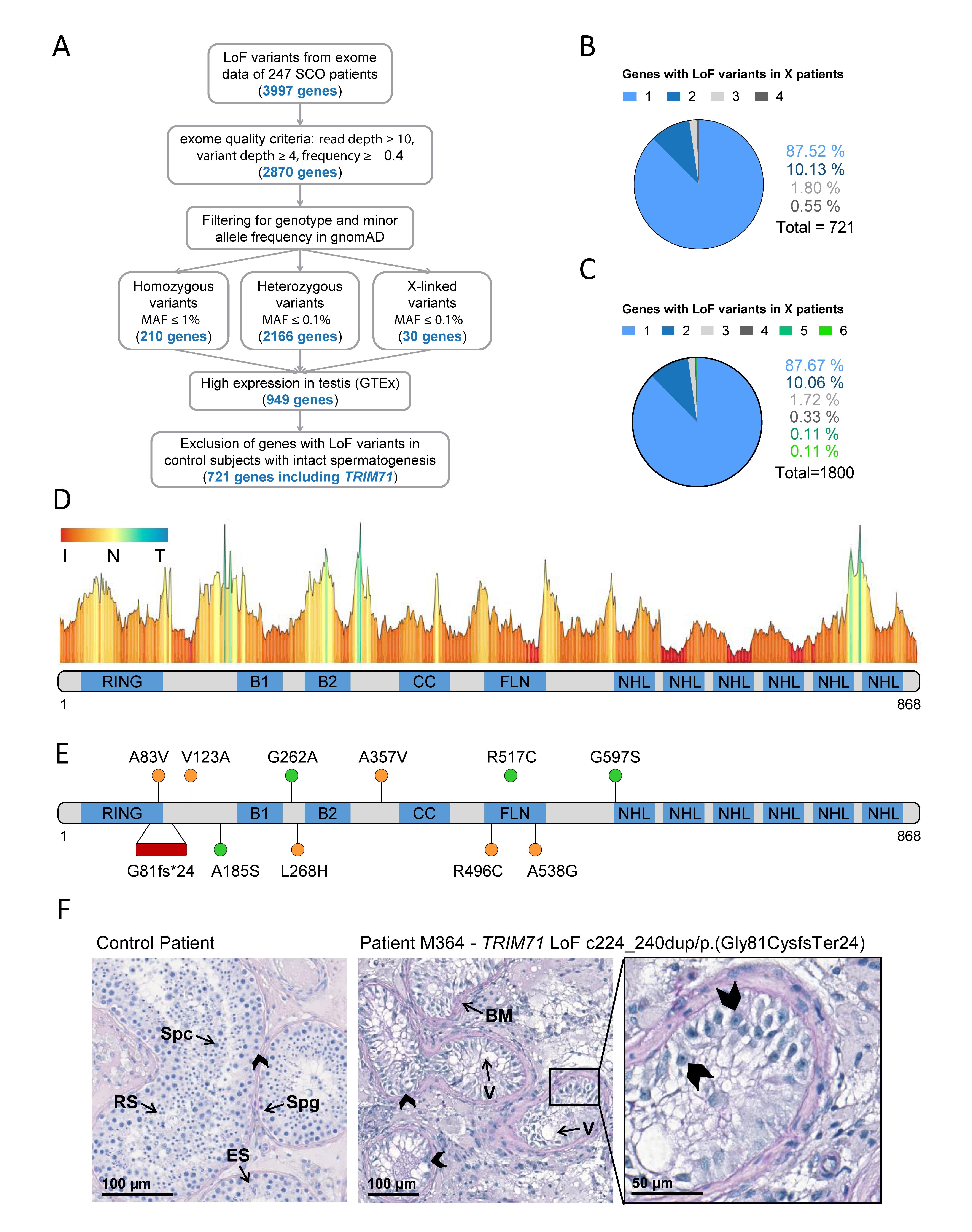
Figure 6. Exome sequencing in infertile SCO patients reveals an association of TRIM71 deficiency with human male infertility.
(A) Filtering scheme detailing gene prioritization of exome sequencing data from 247 individuals with Sertoli cell-only (SCO) phenotype using Sciobase and Haystack (see details in the section “METHODS”). Gene numbers remaining after each filtering step are indicated in blue. Loss-of-function (LoF) variants include stop, frameshift, splice acceptor and splice donor variants. gnomAD, Genome Aggregation Database; MAF, minor allele frequency; GTEx, Genotype-Tissue Expression Portal. (B) Percentages of genes (total = 721, obtained at the end of the filtering process depicted in A) affected by LoF variants in 1–4 SCO patients. None of the genes carried LoF variants in more than 4 patients in the SCO cohort (n = 247). (C) Percentages of genes (total = 1800, obtained at the end of the filtering process depicted in (A), excluding “High expression in testis - GTEx” filtering step) affected by LoF variants in 1-6 SCO patients. None of the genes carried LoF variants in more than 6 patients in the SCO cohort (n = 247). (D) TRIM71 mutation tolerance landscape obtained from MetaDome (https://stuart.radboudumc.nl/metadome/). The graph displays missense over synonymous variant ratios per position for the entire protein, based on gnomAD variants. I, intolerant; N, neutral; T, tolerant. (E) Schematic representation of TRIM71 domain structural organization depicting the location of the genetic variants identified in MERGE. Red, presumably pathogenic LoF variants; orange, missense variants of uncertain significance; green, missense variants found also in proven fathers. (F) Periodic acid–Schiff stainings of testis sections obtained by testicular biopsy from a control patient and an SCO patient (subject M364), who harbors the TRIM71 LoF variant c.224_240dup/p.(Gly81CysfsTer24) in heterozygosis. The control section shows intact spermatogenesis (Spg, spermatogonia; Spc, spermatocytes; RS, round spermatids; ES, elongated spermatids) as opposed to subject M364, whose histological evaluation revealed an SCO phenotype (exemplary Sertoli cells indicated by arrow heads) with several degenerated seminiferous tubules, frequently including large vacuoles (V) and thickened basement membranes (BM). See also Table 1, Supplementary Figures 11, 12, and Supplementary Tables 1, 2.
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 7: Table 1. TRIM71 variants identified in infertile men within the MERGE cohort.
Exome: Whole Exome Sequencing
Species
| Species |
|---|
| human |
Images
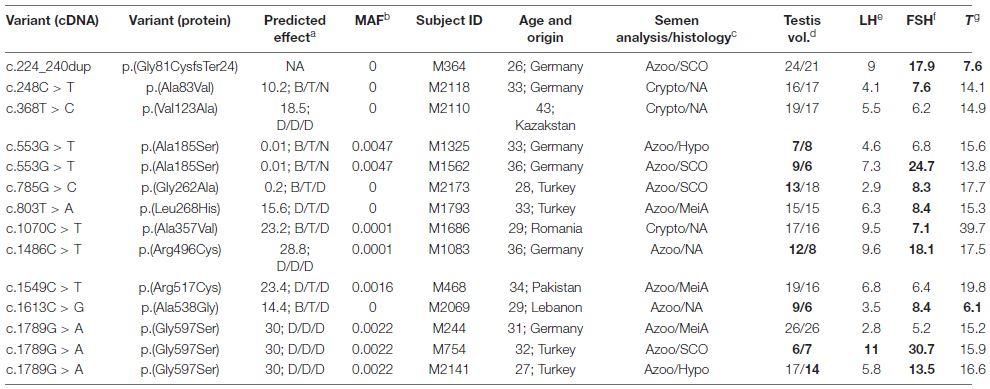
Table 1. TRIM71 variants identified in infertile men within the MERGE cohort.
a) Predicted effect of each TRIM71 variant estimated by four different pathogenicity prediction algorithms (CADD/PolyPhen2/SIFT/MutationTaster, shown in the same order). For CADD, variants with values above 20 are more likely to be deleterious to protein function. For the rest, D, damaging/deleterious; T, tolerated; B, benign; N, neutral; NA, not available. b) MAF (minor allele frequency) values derive from gnomAD and are considered rare if MAF < 0.001 when occurring in heterozygosis. c) Semen analysis was performed for all patients (Azoo, azoospermia; Crypto, cryptozoospermia), and if biopsies were available, patients’ phenotypes were also histologically assessed (SCO, Sertoli cell-only phenotype; MeiA, meiotic arrest; Hypo, hypospermatogenesis). d) Testicular volumes (testis vol., right/left, ref. > 15 mL each; bold, values outside the normal range). e) Luteinizing hormone (LH, ref. 2-10 IU/L; bold, values outside the normal range). f) Follicle-stimulating hormone (FSH, ref. 1-7 IU/L; bold, values outside the normal range). g) Testosterone (T, ref. > 12 nmol/L; bold, values outside the normal range).
Licensed under: https://creativecommons.org/licenses/by/4.0/
