Walk through
The Male Fertility Gene Atlas performs best, if used in a maximized browser window. Some features might not or only partly be visible in a minimized browser window.
Navigation
- Click About to get general information on MFGA.
- Click Publications to get to an overview of available publications.
- Click Search to get to advanced search form.
- Click Walk Through to get to current page.
- Click Contact to get to a contact form and send a message to the MFGA team.
- Click Login to get to login view (login only for system admins).
About
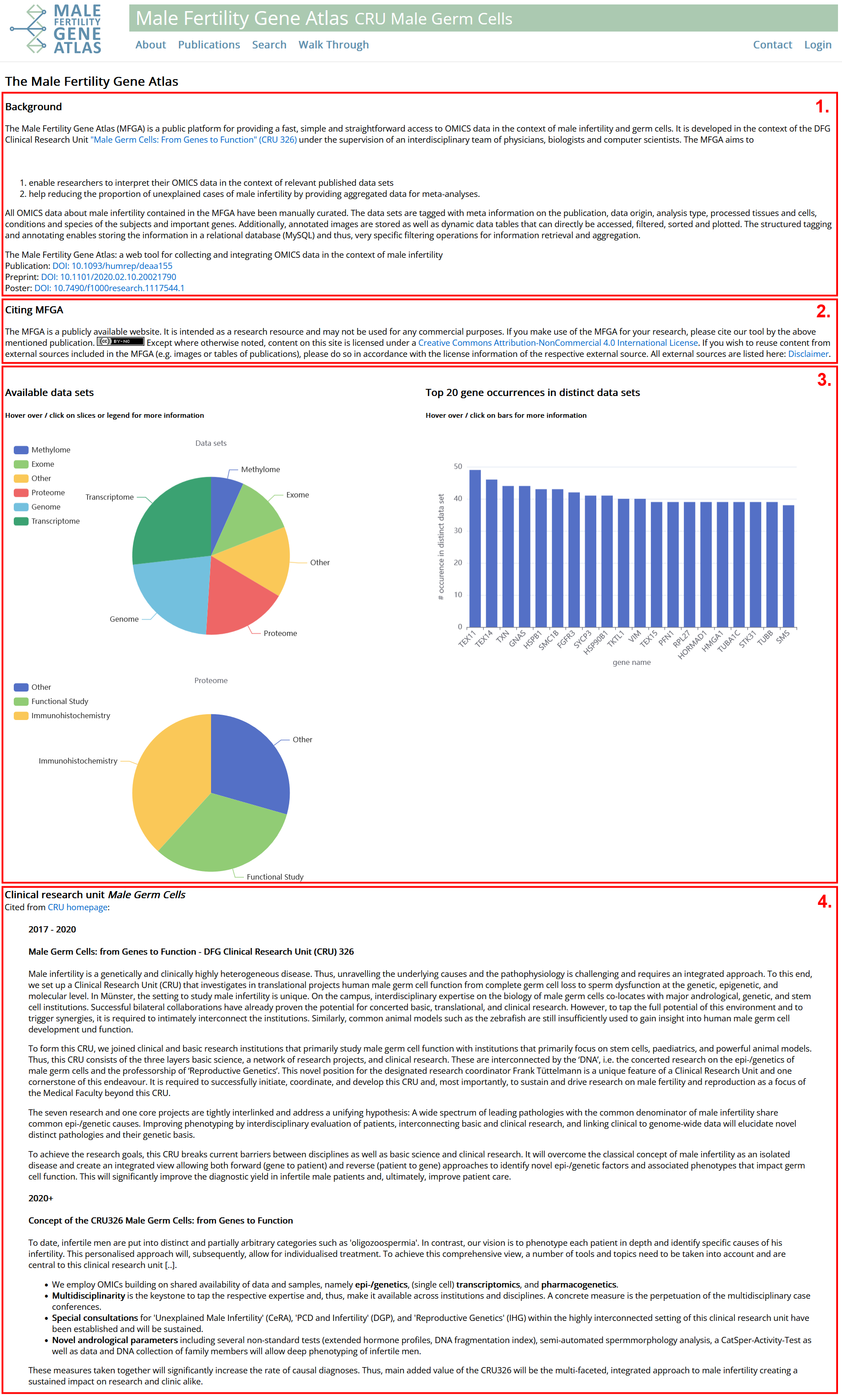
- Information on how MFGA was developed. Links to publication DOI: 10.1093/humrep/deaa155, preprint DOI: 10.1101/2020.02.10.20021790 and poster: DOI: 10.7490/f1000research.1117544.1 with more information on development and specification process.
- Information on how to cite MFGA if used for research.
- Information about data contained in MFGA. Hover over / click on plots to get additional information.
- Information on Clinical Research Unit 326. MFGA is a subproject of this larger research project focused on male infertility.
Publications
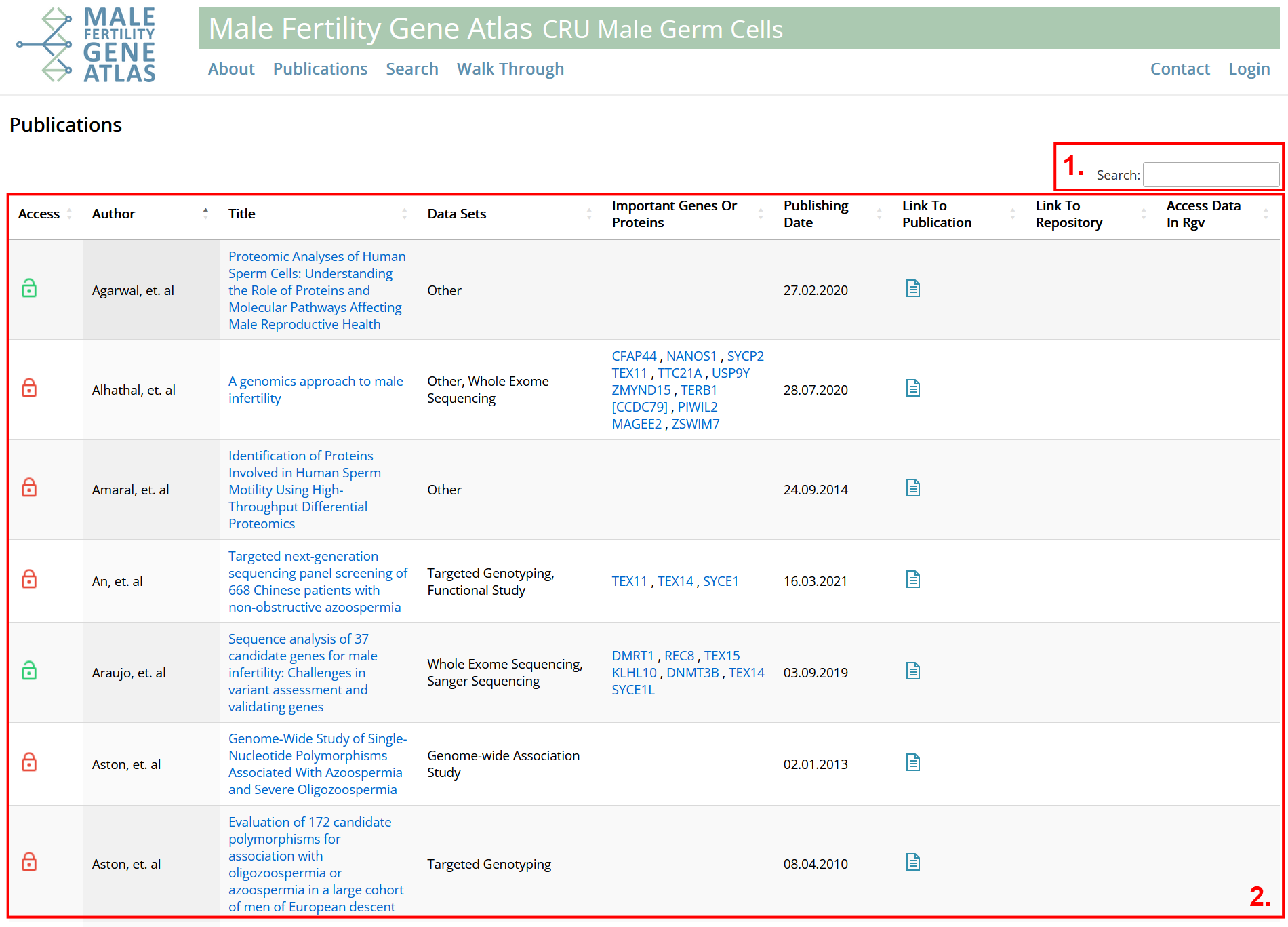
- Simple search form.
- Table with general information on all publications contained in MFGA database. Table can be filtered and sorted column wise. For filtering, enter search text into input fields and press "Enter" or "Tab". Enclosing search text with quotes ("") returns only cells that contain the searched words in given order. Enclosing search text with brackets ([]) returns only cells that match exactly with the search text. For sorting, click buttons next to input fields. Columns left to right:
- Information on how publication is licensed. Green: Fully open access. Yellow: Partly open access. Red: Not open access. See disclaimer for more information.
- Author of publication: Only the first author is shown for better representability. However, filtering works for co-authors as well.
- Title of publication: Click on title to get to a detailed overview of publication.
- Data sets: Categorization of data sets; Curated by MFGA team.
- Important genes or proteins: Genes or proteins emphasized in title or abstract of publication. Click on names to open an overlay with more information and a shortcut to MFGA search for that specific gene or protein.
- Publishing date.
- Link to publication: Click button for redirection to DOI of publication.
- Link to repository: If raw data is provided by the authors, buttons lead to pages where it is published.
- Access data in RGV: If publication is available in RGV, click button for redirection. RGV provides e.g. a genome browser for viewing expression data.
Advanced search
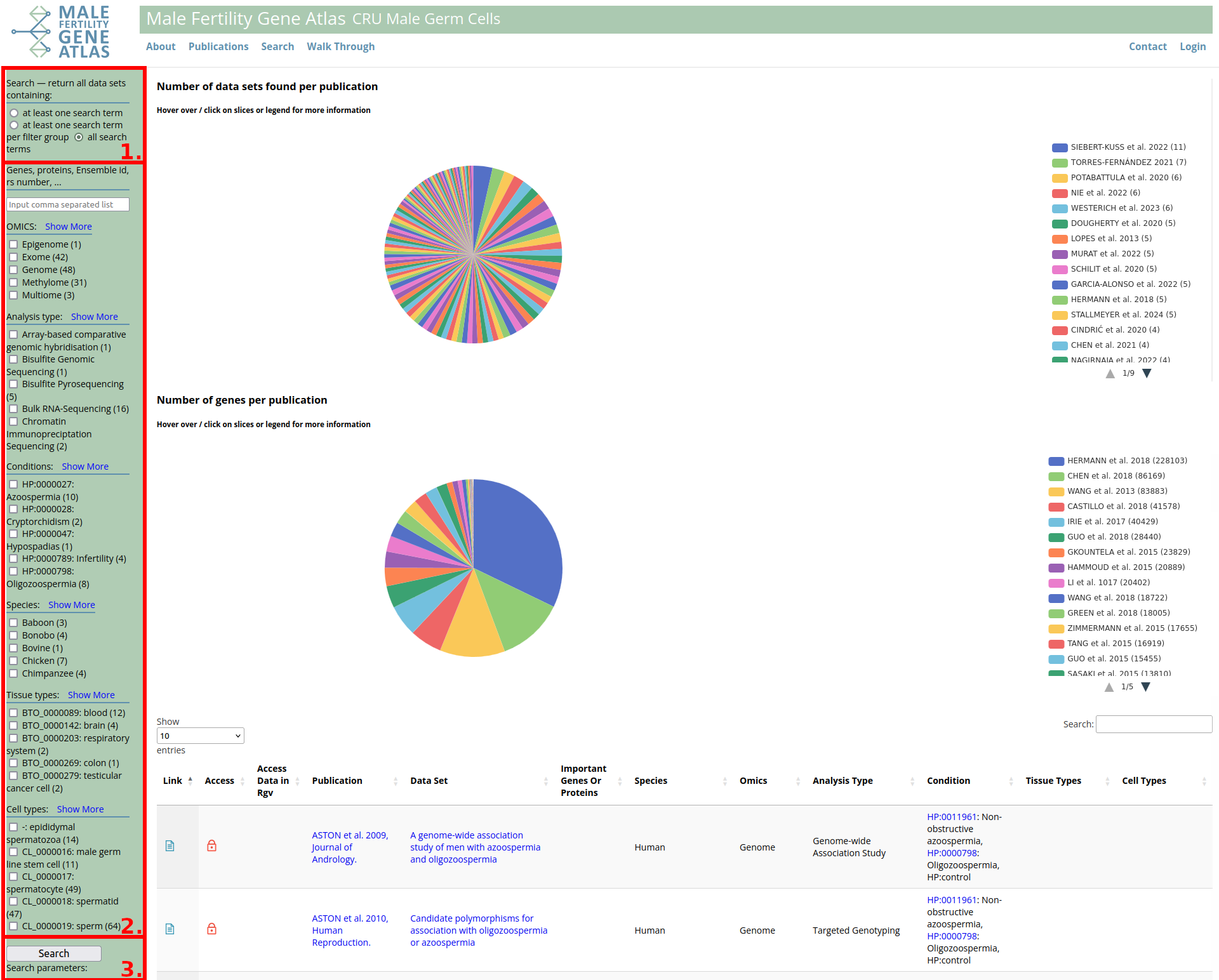
- Search modalities:
- Broad search to identify all data sets that are annotated with at least one of the specified search terms.
- Targeted search for all data sets that are annotated with at least one of the specified search terms per category (gene/ID, OMICS, analysis type, conditions, species, tissue types, cell types).
- Specific search for all data sets that are annotated with each of the search terms (default option).
- Search categories from top to bottom (click on + for more search options):
- Gene/ID: Comma separated list of gene symbols or IDs (e.g. ensembl transcript ID, rs number). Two options for a broader search. Check "Gene symbol: include alias / previous symbols" to include alias symbols and previous symbols of all entered gene symbols to your search. Check "Include ensembl gene / transcript ids <-> gene symbol" to include ensembl gene and transcript IDs of all entered gene symbols and/or include gene symbol that corresponds to entered ensembl gene and transcript IDs. E.g. original search: "TEX11, ENSG00000125398", both options checked and default search modality checked. The search will be executed as "Get all data sets containing information on (TEX11 or TSGA3 or TGC1 or ZIP4 or MZIP4 or Spo22 or ENSG00000120498 or ENST00000374333 or ENSG00000120498 or ENST00000374320 or ENSG00000120498 or ENST00000395889 or ENSG00000120498 or ENST00000344304 or ENSMUSG00000009670 or ENSMUST00000113718 or ENSMUSG00000009670 or ENSMUST00000113716 or ENSMUSG00000009670 or ENSMUST00000009814) and (ENSG00000125398 or SOX9).
- OMICS: Check type of data set you are interested in. Multiple options can be checked. E.g. "Exome" and "Genome". However, each data set is tagged with only one OMICs, such that the search is always executed as "Get all data sets tagged as "Exome" or "Genome".
- Analysis type: Equivalent to OMICS.
- Conditions: Check conditions you are interested in. Conditions are based on Human Phenotype Ontology (HPO) whenever possible, such that they are organised in a tree structure. Click + and - signs to expand or contract tree. Hover over HPO term to see more information or click on it for redirection to corresponding page of HPO project. Conditions in the same tree branch are always queried with logical operator or. Checking a top-level condition automatically expands tree and checks all lower level conditions. Unchecking a lower level condition automatically unchecks all conditions on higher levels.
- Species: Check species you are interested in. E.g. checking "Human" and "Mouse" as well as default search modality returns all data sets comparing human and mouse data. E.g. checking "Human" and "Mouse" as well as one of the other search modalities returns all data sets containing information on humans, or on mice, or on both.
- Tissue types: Check tissue types you are interested in. Tissue types are based on BRENDA tissue ontology (BTO) whenever possible. Hover over BTO term to see more information or click on it for redirection to corresponding page of BTO project. Search options are equivalent to category "species".
- Cell types: Check cell types you are interested in. Cell types are based on cell ontology (CL) whenever possible. Hover over CL term to see more information or click on it for redirection to corresponding page of CL project. Search options are equivalent to category "species".
- Click Search button or press enter to execute search query.
Result of advanced search: Search includes genes/IDs
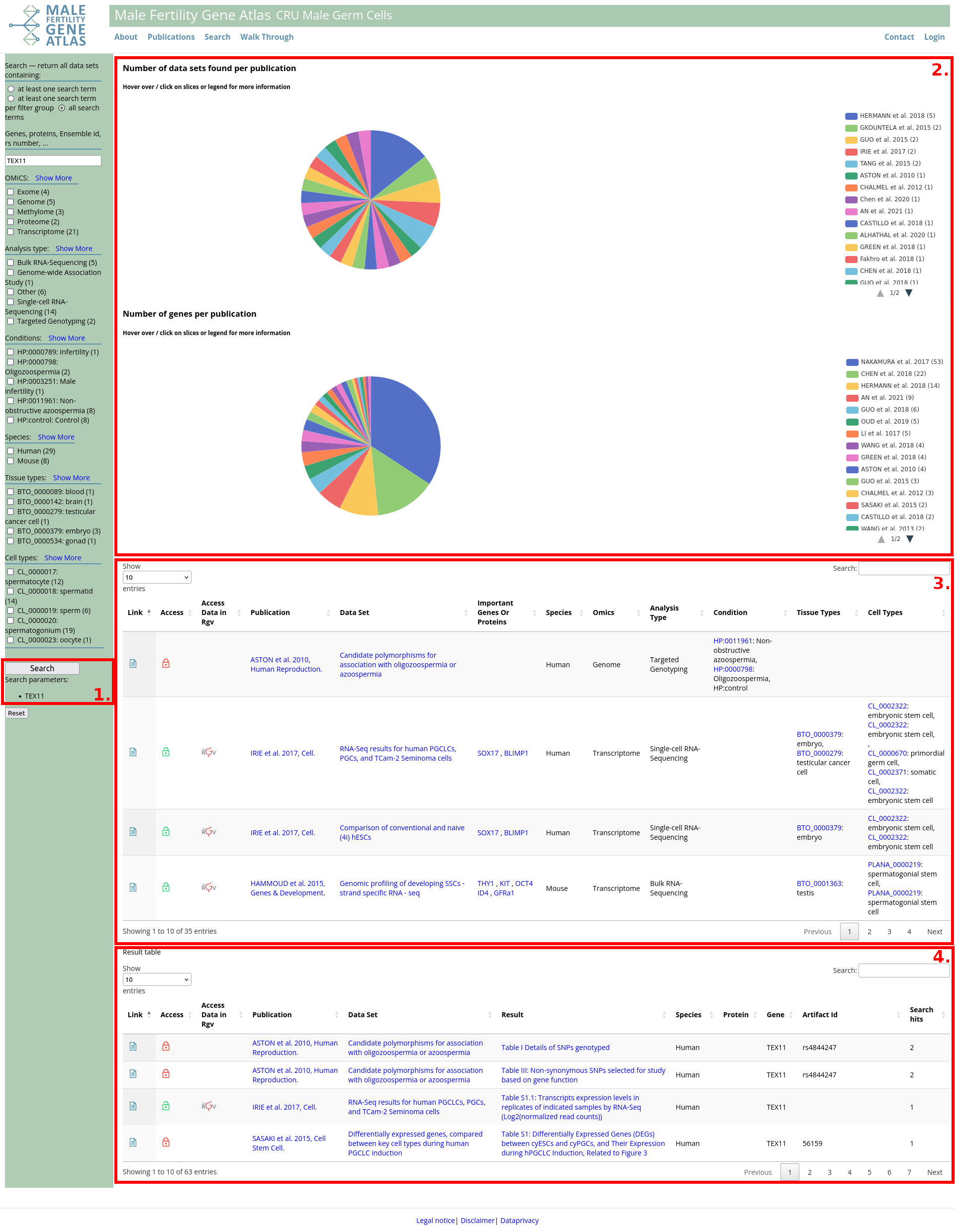
- Search parameters: list of search parameters for current search. Here: Data sets containing information on gene TEX11 and exome data from humans. Search parameters can be reviewed and changed in the advanced search form:.
- Plots: Hover over / click on plots to get additional information.
- Number Of Data Sets Found Per Publication: Data sets matching search request grouped by publication.
- Occurrences Of Genes Per Publication: Number of times genes/IDs from search request occur in data sets grouped by publication.
- Table with data sets matching search request. Structure and filtering /sorting are comparable to section publications. Publication leads to publication overview, data set leads to details on data set in publication overview and result leads to corresponding publication table in MFGA. Filtering this table automatically filters lower table (4.), too.
- Table that lists all occurrences of genes/IDs in tables and images of data sets. Structure and filtering /sorting are comparable to section publications. Rows are grouped by publication / data set / result / gene / ID, such that multiple occurrences of the same gene and ID in one publication table or image are marked in column "Search hits". Click on genes / IDs to get more information.
Publication overview
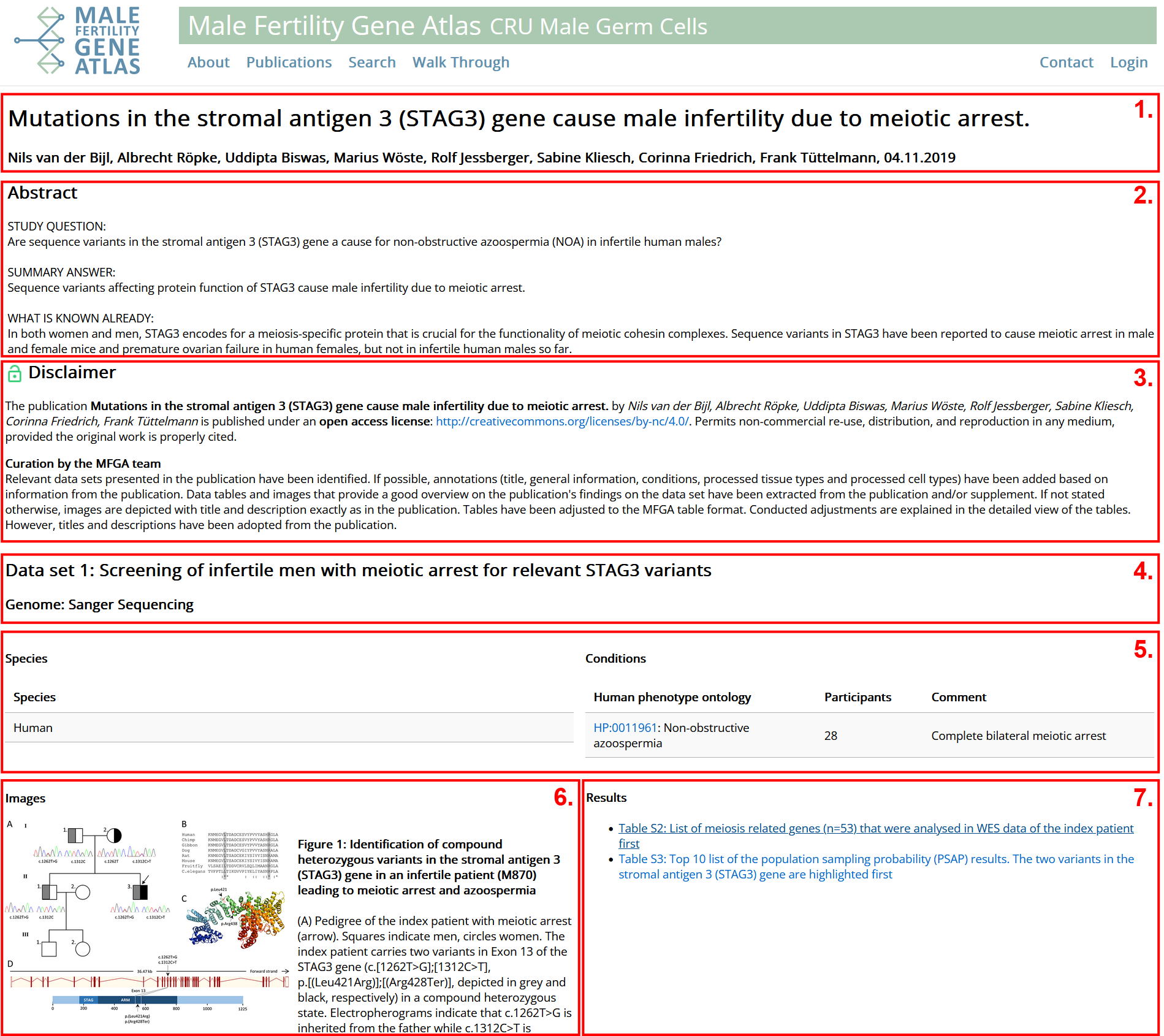
- General Information on publication: title, authors, reference, abstract, link to publication and links to repositories. Abstract is cropped in this example.
- Information on how publication is licensed. Green: Fully open access. Yellow: Partly open access. Red: Not open access. See disclaimer for more information. Depending on specific license, different information can be shown in the overview.
- ata set of publication: Title and type of provided data.
- Species: Species from which data originates.
- Conditions of considered individuals: Conditions are based on Human Phenotype Ontology (HPO) whenever possible.
- Images: Relevant images of data set. Click image to get a larger view. Images are annotated with genes / IDs such that they can be found with a search query.
- Tables: Relevant tables of data set.
Publication table
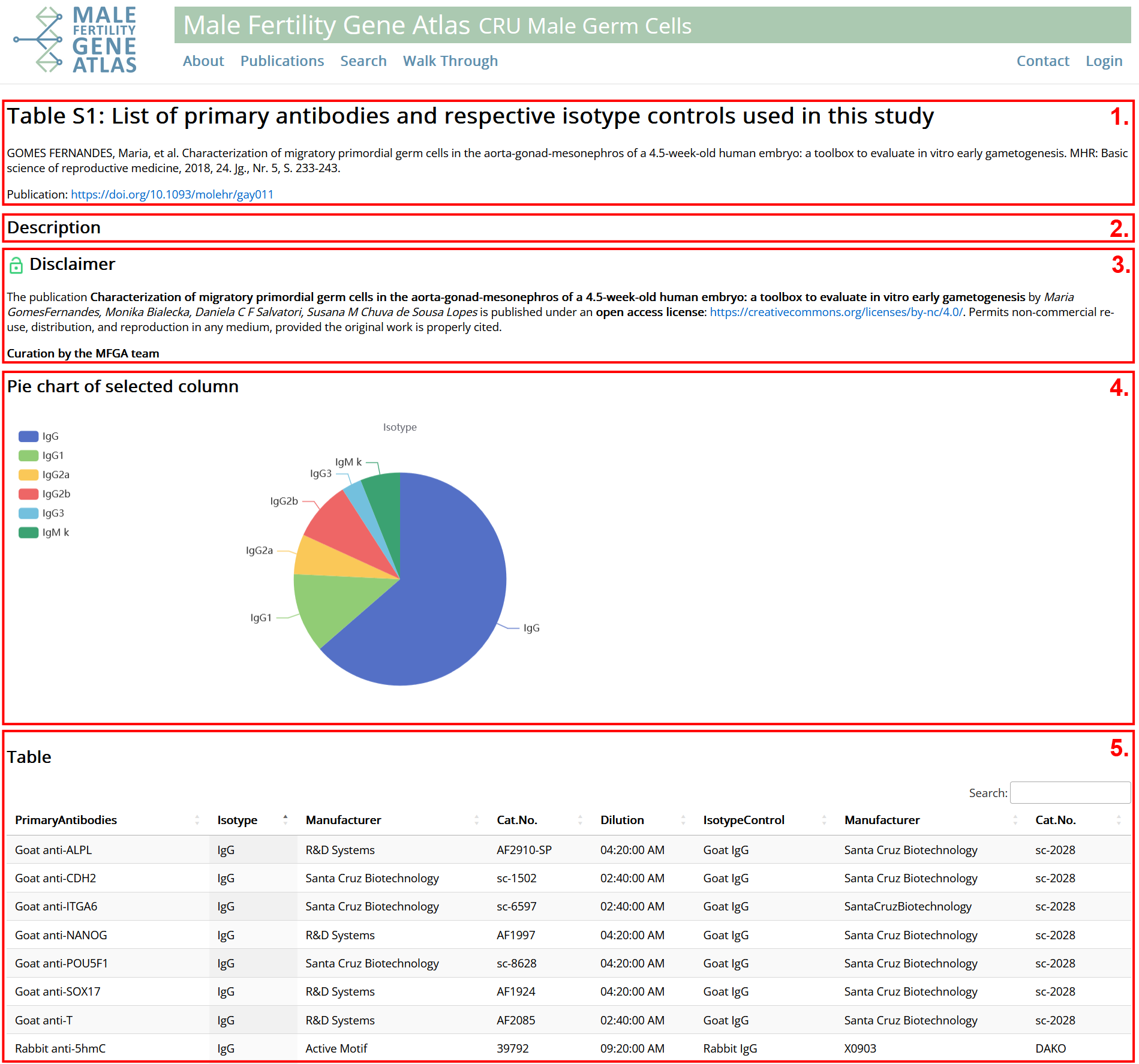
- General information on table: Title, description, reference, link to publication.
- Information on how publication is licensed. Green: Fully open access. Yellow: Partly open access. Red: Not open access. See disclaimer for more information. Tables can only be presented for open access publications.
- Changes on table conducted during manual curation, e.g. renamed / removed columns.
- Plot: Create a histogram on any column of table. Plots: Hover over plot to get additional information. Check "Filtered Table" to adjust plots to current state of filtering of table. Number of breaks can be adjusted for numeric columns. Categorial / text columns can only be represented as histogram if they have at most 100 distinct values.
- Table: As presented in original publication. Structure and filtering /sorting are comparable to section publications.
Overlay
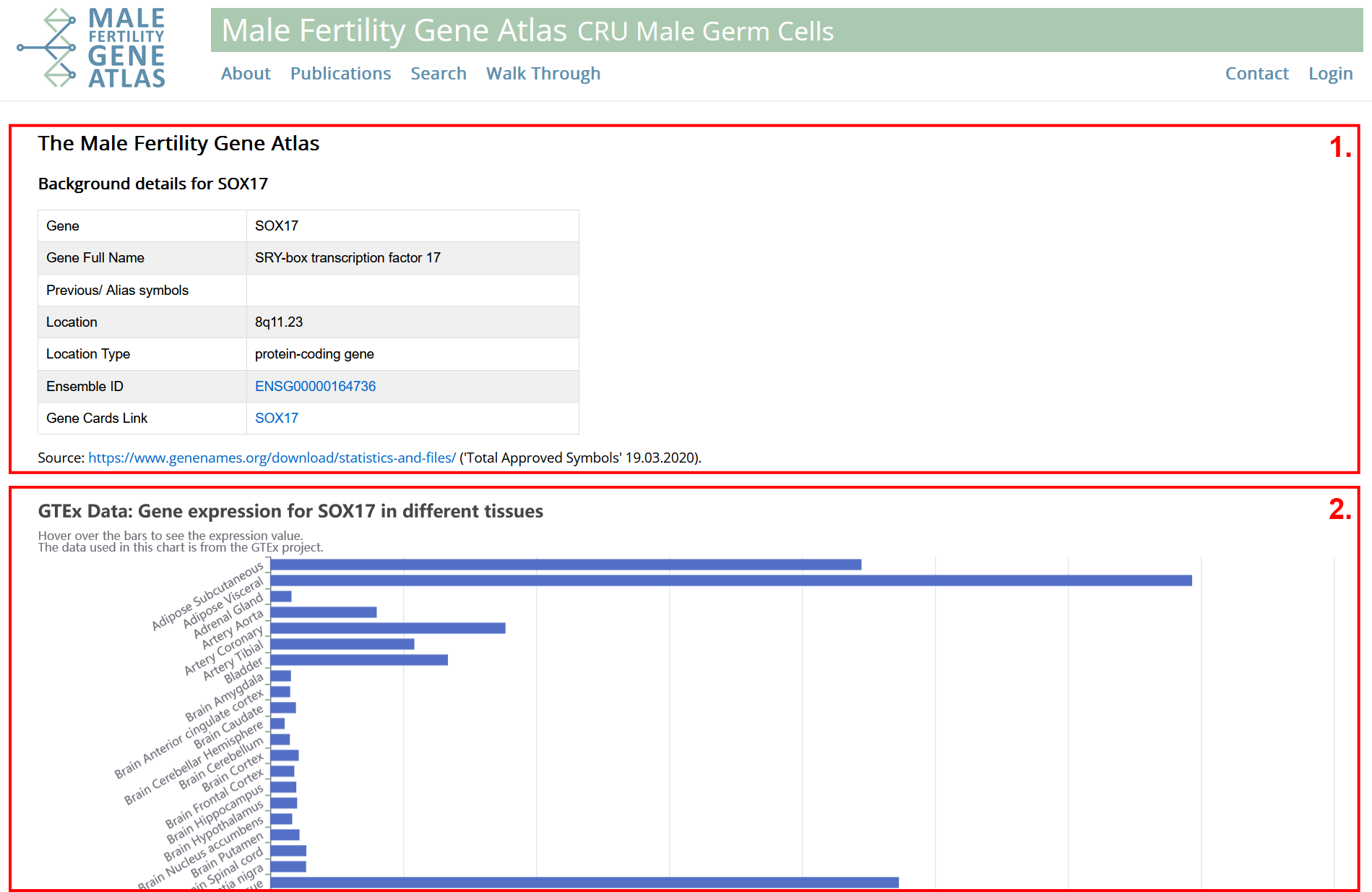
- General information on gene in humans. Data from HGNC. Gene symbols, IDs and proteins are automatically mapped to alias symbols or previous symbols. IDs and proteins are automatically mapped to best matching gene.
- Information on gene expression in different human tissues. Testis tissue is highlighted in red. Data originates from GTEx project.
