Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis
Di-Persio S, Tekath T, Siebert-Kuss LM, Cremers JF, Wistuba J, Li X, Meyer Zu Hörste G, Drexler HCA, Wyrwoll MJ, Tüttelmann F, Dugas M, Kliesch S, Schlatt S, Laurentino S, Neuhaus N, 21.09.2021
Abstract
Despite the high incidence of male infertility, only 30% of infertile men receive a causative diagnosis. To explore the regulatory mechanisms governing human germ cell function in normal and impaired spermatogenesis (crypto), we performed single-cell RNA sequencing (>30,000 cells). We find major alterations in the crypto spermatogonial compartment with increased numbers of the most undifferentiated spermatogonia (PIWIL4+). We also observe a transcriptional switch within the spermatogonial compartment driven by increased and prolonged expression of the transcription factor EGR4. Intriguingly, the EGR4-regulated chromatin-associated transcriptional repressor UTF1 is downregulated at transcriptional and protein levels. This is associated with changes in spermatogonial chromatin structure and fewer Adark spermatogonia, characterized by tightly compacted chromatin and serving as reserve stem cells. These findings suggest that crypto patients are disadvantaged, as fewer cells safeguard their germline's genetic integrity. These identified spermatogonial regulators will be highly interesting targets to uncover genetic causes of male infertility.
Di-Persio S, Tekath T, Siebert-Kuss LM, Cremers JF, Wistuba J, Li X, Meyer Zu Hörste G, Drexler HCA, Wyrwoll MJ, Tüttelmann F, Dugas M, Kliesch S, Schlatt S, Laurentino S, Neuhaus N. Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis. Cell Rep Med. 2021 Sep 9;2(9):100395. doi: 10.1016/j.xcrm.2021.100395. PMID: 34622232; PMCID: PMC8484693.
Publication: https://doi.org/10.1016/j.xcrm.2021.100395 Repository: http://proteomecentral.proteomexchange.org
 Disclaimer
Disclaimer
The publication Single-cell RNA-seq unravels alterations of the human spermatogonial stem cell compartment in patients with impaired spermatogenesis by Di-Persio S, Tekath T, Siebert-Kuss LM, Cremers JF, Wistuba J, Li X, Meyer Zu Hörste G, Drexler HCA, Wyrwoll MJ, Tüttelmann F, Dugas M, Kliesch S, Schlatt S, Laurentino S, Neuhaus N is published under an open access license: https://creativecommons.org/licenses/by-nc-nd/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Alterations of the spermatogonial compartment in cryptozoospermic men
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000020: spermatogonium | An euploid male germ cell of an early stage of spermatogenesis. |
Images
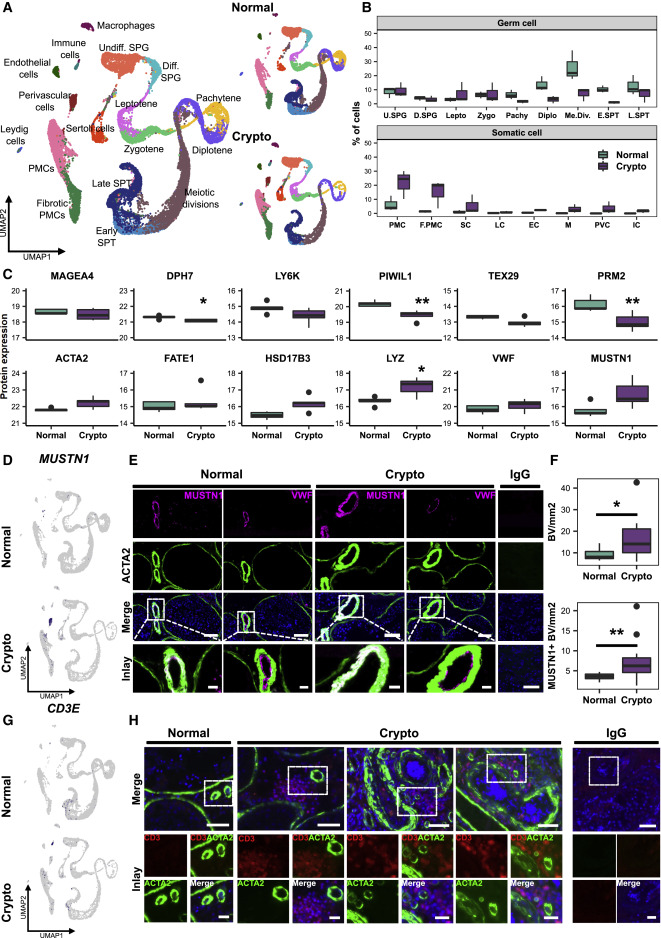
Figure 1. Crypto tissues show stable numbers of MAGEA4+ spermatogonia but reduced numbers of UTF1+ spermatogonia
(A) Experimental outline. (B) Top: representative periodic acid-Schiff stained micrographs of testicular tissue of one normal (left) and one crypto (right) sample. Scale bars, 100 μm. Lower panel: boxplots showing the percentages of tubules containing germ cells (most advanced germ cell type), only Sertoli cells, or tubular shadows in the patient cohort. Values per patient can be found in Table S1. ∗p < 0.05 and ∗∗∗p < 0.001. (C) Micrographs showing the MAGEA4 and UTF1 staining in normal and crypto testicular tissue. Inlays show examples of positive and negative cells for each staining. IgG controls show no staining. Scale bars, 100 μm. (D) Quantification of MAGEA4+ (top) and UTF1+ (lower) spermatogonia per tubule in normal (n = 6) and crypto (n = 6) samples. No statistical difference was found in the number of MAGEA4+ spermatogonia per tubule between normal and crypto. A significant reduction was found in the number of UTF1+ spermatogonia per tubule in the crypto samples (∗p < 0.05; statistical details are available in Table S2). (E) Stacked bar plots showing the ploidy of the single-cell suspensions used for scRNA-seq analysis of each patient sample. (F) Top: uniform manifold approximation and projection (UMAP) plot of the integrated normal dataset. Clusters were assigned on the basis of the expression of 55 marker genes (Figure S1B). Lower panel: scRNA-seq data from the three normal samples. Each color represents a different cell type. (G) Top: UMAP plot of the integrated crypto dataset. The clusters were assigned by anchoring integration using the normal dataset as reference. Lower panel: scRNA-seq data from the three crypto samples.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
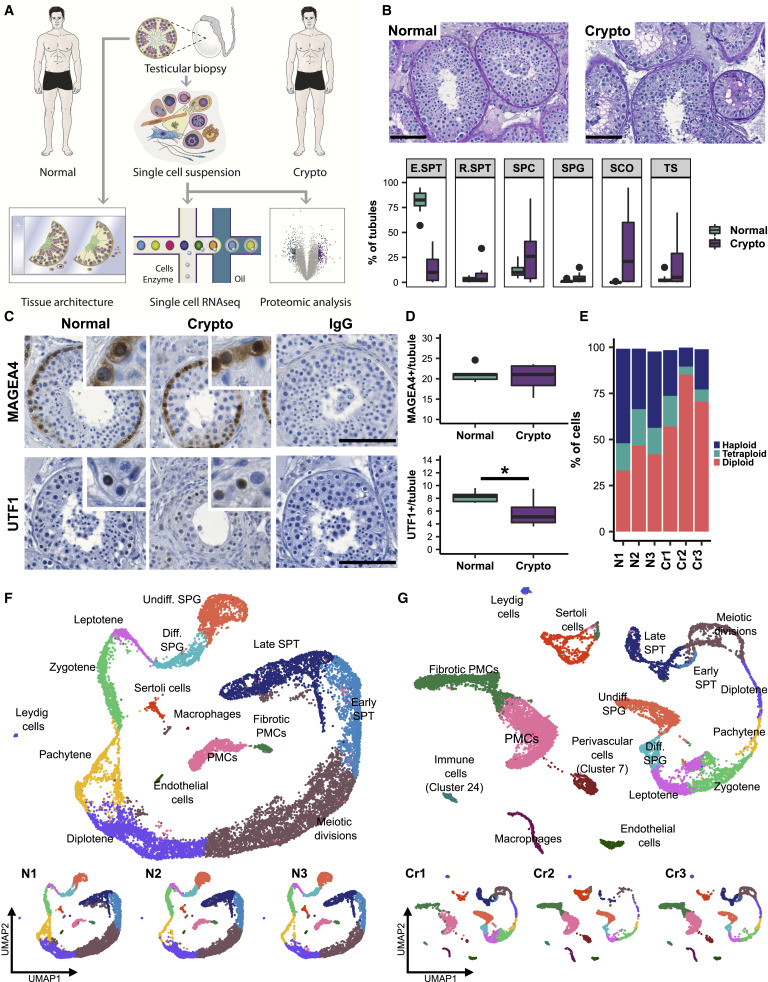
Figure 2. Exploration of cellular and transcriptional changes in crypto testicular tissues
(A) Left panel: UMAP plot of the all integrated dataset. Right panel: contribution of the normal (15,546 cells) and crypto (13,144 cells) datasets to the all integrated dataset. The cells are color coded according to the respective cell types. (B) Boxplot comparing the germ and somatic cell proportions in normal and crypto scRNA-seq datasets. (C) Panel of boxplots representing the protein expression of 12 marker genes used in the cluster assignment (MAGEA4: spermatogonia; DPH7: leptotene spermatocytes; LY6K: zygotene spermatocytes; PIWIL1: pachytene spermatocytes; TEX29: early spermatids; PRM2: elongated spermatids; ACTA2: peritubular myoid cells [PMCs]; FATE1: Sertoli cells; HSD17B3: Leydig cells; VWF: endothelial cells; LYZ: macrophages; MUSTN1: perivascular cells). Protein expression was measured performing bottom-up mass spectrometry analysis on single-cell suspensions (normal n = 5 patients, crypto n = 4 patients) and shows a reduction of germ cells, starting from the leptotene spermatocytes, and an increase of macrophages in crypto patients. ∗p < 0.05 and ∗∗p < 0.01. (D) Feature plots highlighting the expression of MUSTN1 in normal and crypto datasets. (E) Representative micrographs showing blood vessels in consecutive sections of normal and crypto testicular tissues stained for MUSTN1 (magenta)/ACTA2 (green) and for VWF (magenta)/ACTA2 (green). The tissue sections were counterstained with DAPI (blue). Double-positive MUSTN1/ACTA2 cells were found surrounding the endothelial layer of the blood vessels. The IgG control showed no immunological staining. Scale bars, 100 μm (main) and 20 μm (inlays). (F) Boxplots representing the number of ACTA2+ (top) and MUSTN1+ (lower) blood vessels per square millimeter of tissue in normal (n = 12) and crypto samples (n = 13). A significant increase of ACTA2+ and MUSTN1+ blood vessels was found between the two cohorts (∗p < 0.05 and ∗∗p < 0.01; statistical details are available in Table S2). (G) Feature plots highlighting the expression of CD3E (immune cell marker gene) in normal and crypto datasets. (H) Representative micrographs showing CD3+ immune cells (red) and ACTA2+ blood vessels (green) in normal and crypto testicular tissues. The tissue sections were counterstained with DAPI (blue). CD3+ immune cells were found solely in the crypto group in close proximity to blood vessels. The IgG control showed no immunological staining. Scale bars, 50 μm (main) and 20 μm (inlays).
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
