Transient reduction of DNA methylation at the onset of meiosis in male mice
Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A., 04.04.2018
Abstract
Background: Meiosis is a specialized germ cell cycle that generates haploid gametes. In the initial stage of meiosis, meiotic prophase I (MPI), homologous chromosomes pair and recombine. Extensive changes in chromatin in MPI raise an important question concerning the contribution of epigenetic mechanisms such as DNA methylation to meiosis. Interestingly, previous studies concluded that in male mice, genome-wide DNA methylation patters are set in place prior to meiosis and remain constant subsequently. However, no prior studies examined DNA methylation during MPI in a systematic manner necessitating its further investigation. Results: In this study, we used genome-wide bisulfite sequencing to determine DNA methylation of adult mouse spermatocytes at all MPI substages, spermatogonia and haploid sperm. This analysis uncovered transient reduction of DNA methylation (TRDM) of spermatocyte genomes. The genome-wide scope of TRDM, its onset in the meiotic S phase and presence of hemimethylated DNA in MPI are all consistent with a DNA replication-dependent DNA demethylation. Following DNA replication, spermatocytes regain DNA methylation gradually but unevenly, suggesting that key MPI events occur in the context of hemimethylated genome. TRDM also uncovers the prior deficit of DNA methylation of LINE-1 retrotransposons in spermatogonia resulting in their full demethylation during TRDM and likely contributing to the observed mRNA and protein expression of some LINE-1 elements in early MPI. Conclusions: Our results suggest that contrary to the prevailing view, chromosomes exhibit dynamic changes in DNA methylation in MPI. We propose that TRDM facilitates meiotic prophase processes and gamete quality control.
Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A. Transient reduction of DNA methylation at the onset of meiosis in male mice. Epigenetics Chromatin. 2018 Apr 4;11(1):15. doi: 10.1186/s13072-018-0186-0. PMID: 29618374; PMCID: PMC5883305.
Publication: https://doi.org/10.1186/s13072-018-0186-0 Repository: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA326117
 Disclaimer
Disclaimer
The publication Transient reduction of DNA methylation at the onset of meiosis in male mice by Gaysinskaya V, Miller BF, De Luca C, van der Heijden GW, Hansen KD, Bortvin A. is published under an open access license: https://creativecommons.org/licenses/by-nc/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Genome‑wide DNA methylation levels in different cell stages
Methylome: Whole Genome Bisulfite Sequencing
Species
| Species |
|---|
| Mouse |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | Adult | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Mouse |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000017: spermatocyte | Adult | A male germ cell that develops from spermatogonia. The euploid primary spermatocytes undergo meiosis and give rise to the haploid secondary spermatocytes which in turn give rise to spermatids. | Mouse | ||
| CL_0000020: spermatogonium | Adult | An euploid male germ cell of an early stage of spermatogenesis. | Mouse | ||
| CL_0000018: spermatid | Adult | A male germ cell that develops from the haploid secondary spermatocytes. Without further division, spermatids undergo structural changes and give rise to spermatozoa. | Mouse | ||
| -: epididymal spermatozoa | Adult | epididymal spermatozoa | Mouse |
Images
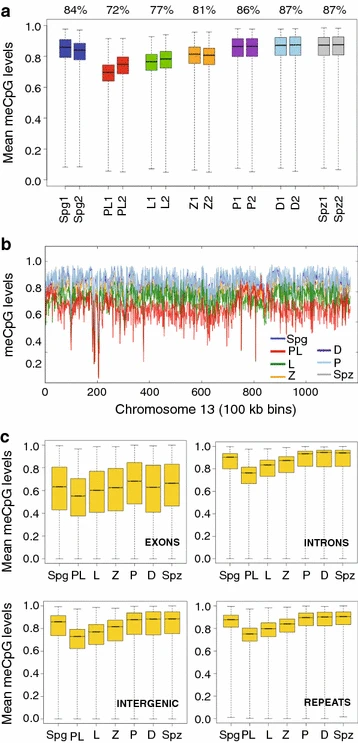
Figure 1: Global DNA methylation dynamics in MPI
a Genome-wide DNA methylation was summarized as means of non-overlapping bins of 500 CpGs for individual biological replicates. Box-and-whisker plot shows the maximum, upper quartile, median, lower quartile and minimum of data. Median percent DNA methylation for both replicates is specified above the boxplot. b Chromosome-wide DNA methylation levels were plotted across chromosome length (chromosome 13, replicate 1 is shown). DNA methylation was averaged using sliding non-overlapping bins of 100 kbp. c Box-and-whisker plot of DNA methylation levels across various genomic features. The average DNA methylation levels were aggregated as consecutive, non-overlapping averages of 100 CpGs. Averages were combined for biological replicates
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
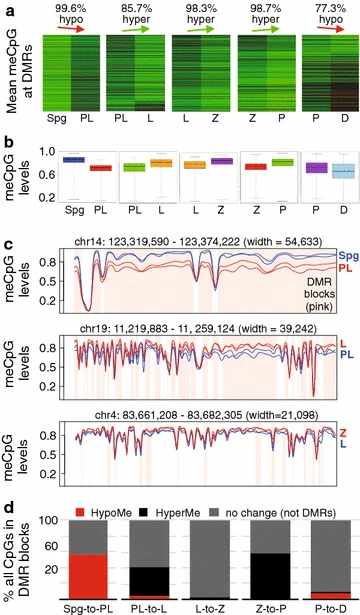
Figure 2: DMR dynamics in MPI
a Heatmap of DNA methylation profiles of all DMRs between consecutive MPI substages. Each row shows DNA methylation level of a DMR in a pairwise comparison. DNA methylation level is scaled according to red (low)-to-green (high) color scale. The percentage of DMRs exhibiting the main direction of DNA methylation change is indicated above the plots. b Boxplot showing DNA methylation value distribution at DMRs in MPI between two consecutive stages. c Smoothed DNA methylation at DMRs for Spg and PL (top), PL and L (middle), and P and D (bottom). d The proportion of CpGs accounted for by hypomethylated and hypermethylated DMRs
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/

Figure 3: DNA methylation pattern in PL overlaps with replication timing
a The plot of CpG DNA methylation averaged using sliding non-overlapping 100-kbp windows in Spg, PL and L across a region of chromosome 14 (top) and replication timing (RT) data for the same region of chromosome 14 from mouse B cell lymphoma CH12 cells (bottom). b Normalized genome sequencing coverage after WGBS summarized as averages of sliding non-overlapping 5-kbp windows
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
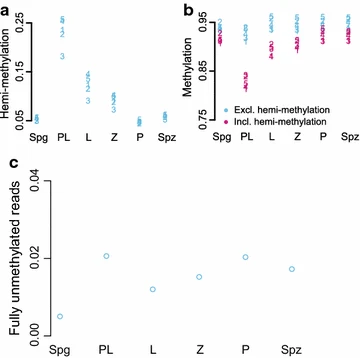
Figure 4: Hemimethylation of L1. Hemimethylated, methylated and unmethylated levels were quantified at 5 CpGs in L1
a The proportion of reads supporting hemimethylation at each of the 5 CpGs in six different cell states. b The amount of methylation at the 5 CpGs quantified including hemimethylation (pink) and excluding hemimethylation (blue). c Levels of fully unmethylated L1 elements in MPI based on reads where all 5 CpGs are unmethylated on both strands
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
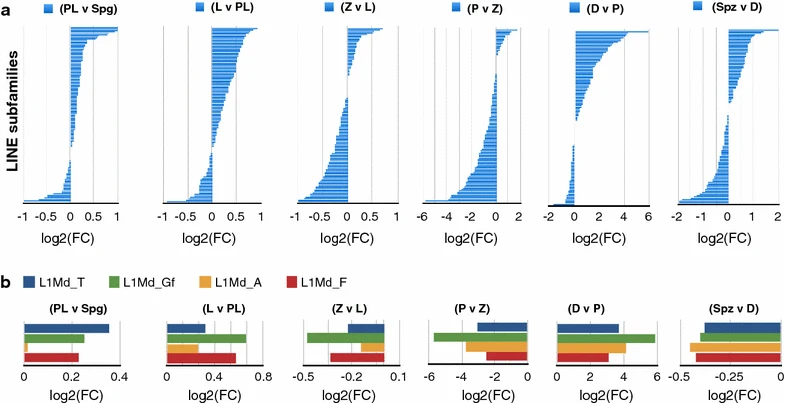
Figure 5: Dynamics of LINE transcript abundance in MPI
a, b A pairwise differential expression analysis is represented as log2 fold change in CPM between consecutive MPI stages and also sperm (Spz) relative to diplotene stage. Each horizontal barplot shows log2(FC) on the x-axis for a different LINE subfamily on the y-axis or b LINE-1 subfamilies that contain evolutionarily young and potentially active members
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
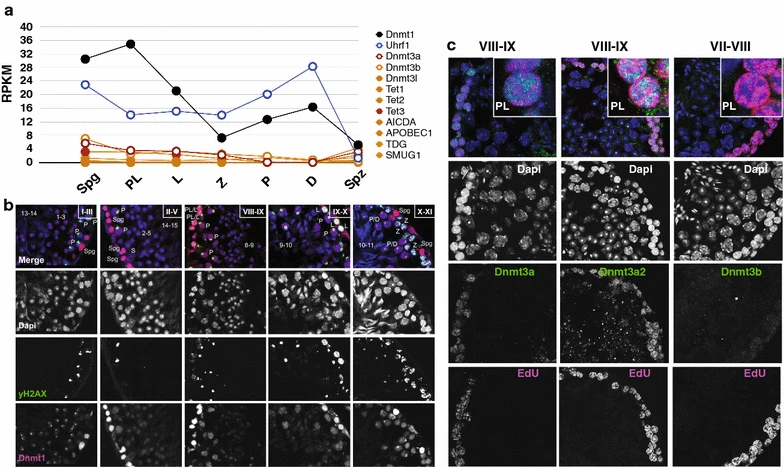
Figure 6: Examination of transcript and protein abundance of genes associated with passive or active DNA demethylation or remethylation
a RNA-seq transcript abundance of select genes in individual FACS-enriched MPI germ cells, expressed as RPKM. b Immunofluorescence co-staining of testicular cross sections for Dnmt1 (pink) and yH2AX (green), and counterstaining with DAPI (blue). The Roman numerals denote spermatogenic stages; the numbers correspond to steps of haploid sperm development during spermatogenesis. Spermatogonia (Spg), preleptonema (PL), leptonema (L) and zygonema (Z), pachynema (P), diplonema (D), Sertoli cells (S). c Immunofluorescence staining of testicular cross sections (cryosections) for EdU (pink) and Dnmt3a, Dnmt3a2 or Dnmt3b (green), and counterstaining with DAPI (blue)
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
