Single-cell roadmap of human gonadal development
Garcia-Alonso L, Lorenzi V, Mazzeo CI, Alves-Lopes JP, Roberts K, Sancho-Serra C, Engelbert J, Marečková M, Gruhn WH, Botting RA, Li T, Crespo B, van Dongen S, Kiselev VY, Prigmore E, Herbert M, Moffett A, Chédotal A, Bayraktar OA, Surani A, Haniffa M, Vento-Tormo R, 06.07.2022
Abstract
Gonadal development is a complex process that involves sex determination followed by divergent maturation into either testes or ovaries1. Historically, limited tissue accessibility, a lack of reliable in vitro models and critical differences between humans and mice have hampered our knowledge of human gonadogenesis, despite its importance in gonadal conditions and infertility. Here, we generated a comprehensive map of first- and second-trimester human gonads using a combination of single-cell and spatial transcriptomics, chromatin accessibility assays and fluorescent microscopy. We extracted human-specific regulatory programmes that control the development of germline and somatic cell lineages by profiling equivalent developmental stages in mice. In both species, we define the somatic cell states present at the time of sex specification, including the bipotent early supporting population that, in males, upregulates the testis-determining factor SRY and sPAX8s, a gonadal lineage located at the gonadal-mesonephric interface. In females, we resolve the cellular and molecular events that give rise to the first and second waves of granulosa cells that compartmentalize the developing ovary to modulate germ cell differentiation. In males, we identify human SIGLEC15+ and TREM2+ fetal testicular macrophages, which signal to somatic cells outside and inside the developing testis cords, respectively. This study provides a comprehensive spatiotemporal map of human and mouse gonadal differentiation, which can guide in vitro gonadogenesis.
Garcia-Alonso L, Lorenzi V, Mazzeo CI, Alves-Lopes JP, Roberts K, Sancho-Serra C, Engelbert J, Marečková M, Gruhn WH, Botting RA, Li T, Crespo B, van Dongen S, Kiselev VY, Prigmore E, Herbert M, Moffett A, Chédotal A, Bayraktar OA, Surani A, Haniffa M, Vento-Tormo R. Single-cell roadmap of human gonadal development. Nature. 2022 Jul;607(7919):540-547. doi: 10.1038/s41586-022-04918-4. Epub 2022 Jul 6. PMID: 35794482; PMCID: PMC9300467.
Publication: https://doi.org/10.1038/s41586-022-04918-4
 Disclaimer
Disclaimer
The publication Single-cell roadmap of human gonadal development by Garcia-Alonso L, Lorenzi V, Mazzeo CI, Alves-Lopes JP, Roberts K, Sancho-Serra C, Engelbert J, Marečková M, Gruhn WH, Botting RA, Li T, Crespo B, van Dongen S, Kiselev VY, Prigmore E, Herbert M, Moffett A, Chédotal A, Bayraktar OA, Surani A, Haniffa M, Vento-Tormo R is published under an open access license: https://creativecommons.org/licenses/by-nc/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Human–mouse single-cell atlases of gonadal and extragonadal tissue
Multiome: Other
Species
| Species |
|---|
| Human |
| Mouse |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | first and second trimesters of gestation (6–21 PCW); at embryonic days (E) 10.5, 11.5 and 12.5 | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human, Mouse | 22 |
| BTO_0000975: ovary | first and second trimesters of gestation (6–21 PCW); at embryonic days (E) 10.5, 11.5 and 12.5 | One of the typically paired essential female reproductive organs that produce eggs and in vertebrates female sex hormones. | Human, Mouse | 33 |
Images
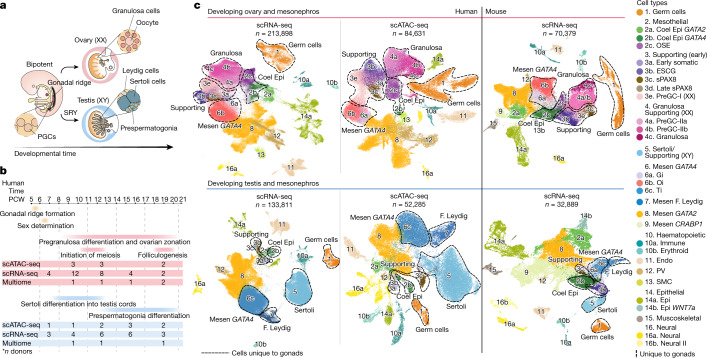
Figure 1: Human–mouse harmonized single-cell atlases of gonadal and extragonadal tissue
a, Schematic illustration of gonadal development showing the main structures of the XX and XY gonads. b, Diagram summarizing the stage and sex composition of our sample cohort along with main events occurring during gonadogenesis. c, Top shows the UMAP of cell lineages (colour) in the human female scRNA-seq (n = 213,898), human female scATAC-seq (n = 84,631) and mouse female scRNA-seq (n = 70,379) datasets. Bottom shows UMAP projections of cell lineages (colour) in the human male scRNA-seq (n = 133,811), human male scATAC-seq (n = 52,285) and mouse male scRNA-seq (n = 32,889) datasets. Clusters for mesothelial, supporting and gonadal mesenchymal LHX9+ cells were defined in an independent per-lineage reanalysis and projected onto this dataset (Fig. (Fig.3).3). Dashed lines outline the cell populations unique to the gonads. Doublets and low-quality control cells were removed. CoelEpi, coelomic epithelium; Endo, endothelial; Epi, epithelial; F. Leydig, fetal Leydig; Gi, gonadal interstitial; Mesen, mesenchymal; Oi, ovarian interstitial; OSE, ovarian surface epithelium; preGC, pregranulosa cells; PV, perivascular; sPAX8, supporting PAX8 +; Ti, testicular interstitial; SMC, smooth muscle cell.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 2: TFs modulating germ cell differentiation
Multiome: Other
Species
| Species |
|---|
| Human |
| Mouse |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000975: ovary | One of the typically paired essential female reproductive organs that produce eggs and in vertebrates female sex hormones. | Human, Mouse | ||
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human, Mouse |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000586: germ cell | The reproductive cell in multicellular organisms | Human, Mouse |
Images
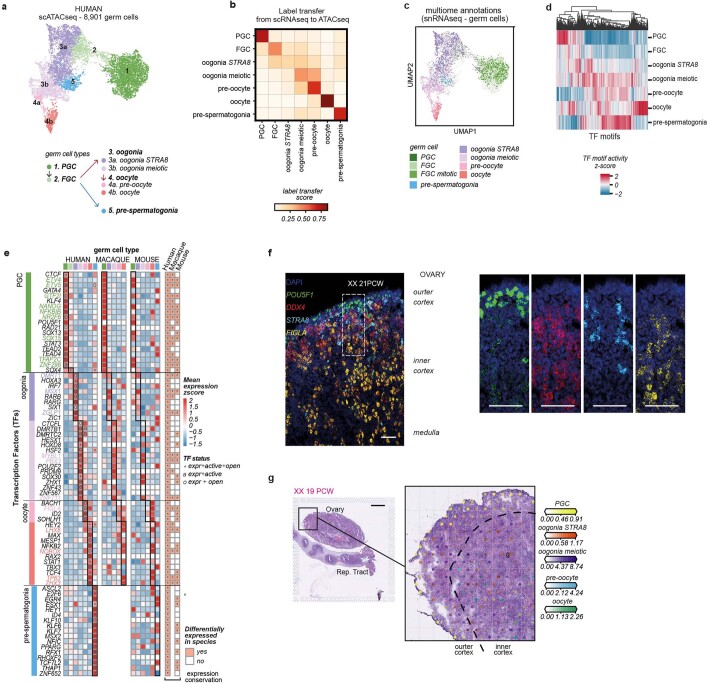
Figure 2: Cross species TF comparison of germ cells
a, UMAP of germ cell states (colour) in the human scATAC-seq (n = 8,901) dataset. Doublets and low QC cells removed. b, Heatmap reporting label transfer scores from human scRNA-seq to scATAC-seq germ cell data of matched individuals. c, UMAP of germ cells in the human scATAC-seq dataset labelled by cell state identified in snRNA-seq data from the cell-coupled snRNA-seq/snATAC-seq profiling. d, Hierarchical clustering of transcription factor (TF) binding activity scores in each human germ cell type estimated from scATAC-seq data. e, Heatmaps showing the expression of human-relevant transcription factors (TF) in human, macaque and mouse germ cells. Colour proportional to scaled log-transformed expression. For human germ cells only: “o” = TF whose binding motifs are differentially accessible (i.e. TF can bind their potential targets); “a” = TF whose targets are differentially expressed (i.e. differentially activated TF); and asterisk (*) = TF that meets both “o” and “a” conditions. Conservation heatmap (right) highlights significant overexpression (log2-fold change > 0 and FDR < 0.05) in each species. TFs whose upregulation is conserved across species are highlighted with bold/coloured labels. f, High-resolution imaging of a representative transverse section of a human ovary at 21 post-conceptional weeks (PCW), with intensity proportional to smFISH signal for POU5F1 (green, primordial germ cells), DDX4 (red, fetal germ cells), STRA8 (cyan, pre-meiotic germ cells) and FIGLA (yellow, oocytes); n = 4. The white dashed rectangle highlights the enlarged gonadal region. Scale bars = 100 µm. g, cell2location estimated cell abundance (colour intensity) contributed by each germ cell to each Visium spot (colour) shown over the H&E image of a 19 PCW ovary; n = 2. Scale bars = 1 mm. E = embryonic day; Expr = expressed, FGC = fetal germ cells; P = postnatal day; PCW = post-conceptional weeks; PGC = primordial germ cells.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 3: New gonadal somatic cells during sex determination
Multiome: Other
Species
| Species |
|---|
| Human |
| Mouse |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human, Mouse | ||
| BTO_0000975: ovary | One of the typically paired essential female reproductive organs that produce eggs and in vertebrates female sex hormones. | Human, Mouse |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| PLANA_0002096: somatic gonadal cell | A non-germline cell within the ovary or testis. | Human, Mouse | |||
| CL_0000501: granulosa cell | A supporting cell for the developing female gamete in the ovary of mammals. They develop from the coelomic epithelial cells of the gonadal ridge. Granulosa cells form a single layer around the mammalian oocyte in the primordial ovarian follicle and advance to form a multilayered cumulus oophorus surrounding the ovum in the Graafian follicle. The major functions of granulosa cells include the production of steroids and LH receptors. | Human, Mouse | |||
| CL_0000216: Sertoli cell | A supporting cell projecting inward from the basement membrane of seminiferous tubules. They surround and nourish the developing male germ cells and secrete androgen binding protein. Their tight junctions with the spermatogonia and spermatocytes provide a blood-testis barrier. | Human, Mouse |
Images
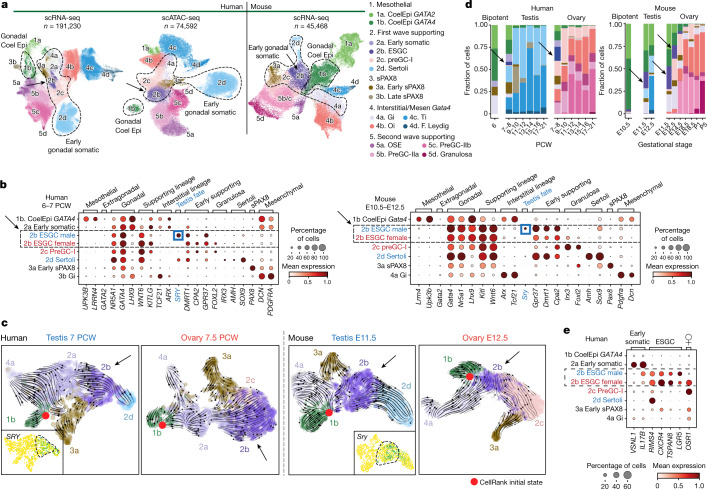
Figure 3: New gonadal somatic cells during sex determination in humans and mice
a, UMAP of somatic cell states (colour) in the human scRNA-seq (n = 191,230), human scATAC-seq (n = 74,592) and mouse scRNA-seq (n = 45,468) datasets. Doublets and low-quality control cells were removed. b, Dot plots show the variance-scaled, log-transformed expression of genes (x-axis) characteristic of the first wave of somatic cells (y-axis) in humans and mice. c, UMAP of somatic cells overlaid with RNA velocity maps in two humans (7 PCW testis; 7.5 PCW ovary) and two mice (E11.5 testis, E12.5 ovary) gonadal samples, analysed independently. d, Relative proportions of human and mouse somatic cell states (colour) profiled with scRNA-seq, classified by sex and developmental stage. Black arrows highlight the ESGCs. e, Dot plot showing the variance-scaled, log-transformed expression of human-specific early somatic and ESGC markers (x-axis) in the first wave of human supporting cells (y-axis). CoelEpi, coelomic epithelium; Gi, gonadal interstitial; Oi, ovarian interstitial; preGC, pregranulosa cells; sPAX8, supporting PAX8; Ti, testicular interstitial.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 4: Supporting-like PAX8+ (sPAX8) gonadal lineage forms the rete testis
Other: smFISH
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | ||
| BTO_0000975: ovary | One of the typically paired essential female reproductive organs that produce eggs and in vertebrates female sex hormones. | Human |
Images
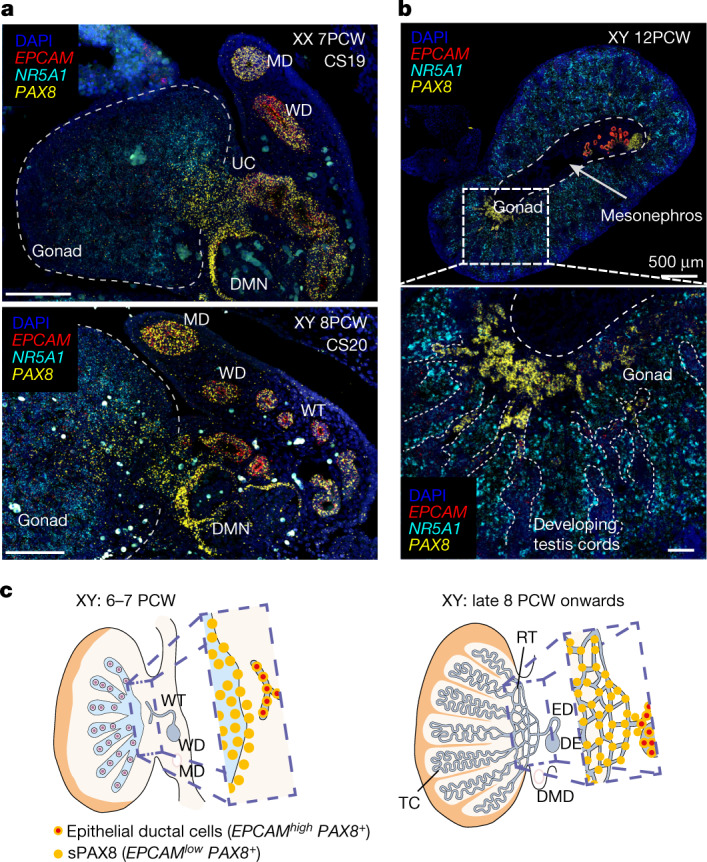
Figure 4: Supporting-like PAX8+ (sPAX8) gonadal lineage forms the rete testis
a, High-resolution large-area imaging of representative gonadal sections (transverse) of a human ovary (7 PCW, CS19; top) and testis (8 PCW, CS20; bottom), with intensity proportional to smFISH signal for EPCAM (red, epithelial), NR5A1 (cyan, gonadal somatic) and PAX8 (yellow, sPAX8 and epithelial) (n = 2); red blood cells appear as bright autofluorescent cells. b, High-resolution large-area imaging of representative gonadal sections of one human testis (12 PCW, transverse section), with intensity proportional to smFISH signal for EPCAM (red, epithelial), NR5A1 (cyan, gonadal somatic) and PAX8 (yellow, sPAX8 and epithelial) (n = 2). White dashed rectangles highlight enlarged gonadal regions with PAX8high EPCAMlow expression. c, Schematic representation of sPAX8 cells in the human testis at two developmental stages. DE, ductus epididymidis; DMD, degenerating Müllerian duct; DMN, degenerating mesonephric nephron; ED, efferent ductule; MD, Mullerian duct; RT, rete testis; TC, testis cords; UC, urogenital connection; WD, Wolffian duct; WT, Wolffian tubules; scale bars, 100 µm unless otherwise specified.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 5: Transcriptional, spatiotemporal and paracrine signatures of human pregranulosa cells
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
| Mouse |
| Macaque |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000975: ovary | One of the typically paired essential female reproductive organs that produce eggs and in vertebrates female sex hormones. | Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000501: granulosa cell | A supporting cell for the developing female gamete in the ovary of mammals. They develop from the coelomic epithelial cells of the gonadal ridge. Granulosa cells form a single layer around the mammalian oocyte in the primordial ovarian follicle and advance to form a multilayered cumulus oophorus surrounding the ovum in the Graafian follicle. The major functions of granulosa cells include the production of steroids and LH receptors. | Human, Mouse |
Images
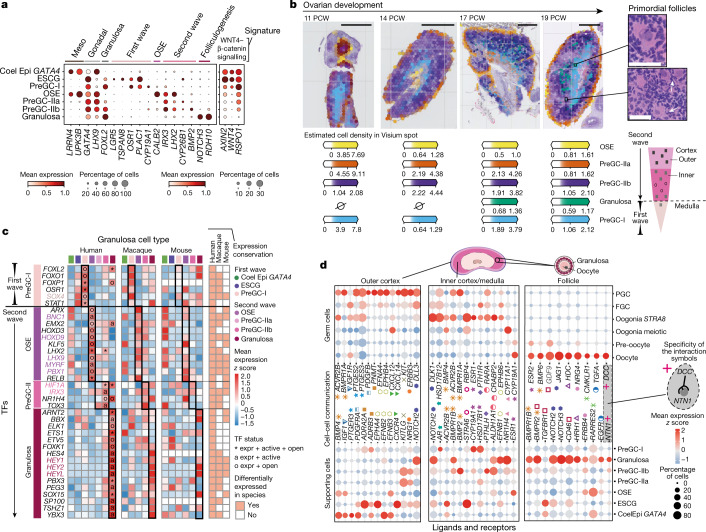
Figure 5: Transcriptional, spatiotemporal and paracrine signatures of human pregranulosa cells
a, Dot plots show the variance-scaled, log-transformed expression of genes (x-axis) characteristic of ovarian supporting cells (y-axis) in human scRNA-seq data. Top layer groups marker genes by categories. b, Spatial mapping of granulosa cell types from the scRNA-seq human dataset to spatial transcriptomics slide of 11, 14, 17 and 19 PCW ovaries using cell2location; n = 2. Estimated cell abundance (colour intensity) for OSE, preGC-I, preGC-IIa, preGC-IIb and developing granulosa cells (colour) in each Visium spot shown over the haematoxylin and eosin (H&E) images. The black rectangles highlight enlarged ovarian regions with forming follicles (top right). Schematic representation of the spatial organization of pregranulosa cell states in the human ovary (bottom right). Scale bars 1 mm (left) and 50 µm in magnified regions (right). c, Heatmaps showing expression of selected TFs across human, macaque and mouse ovarian supporting cells. Colour proportional to scaled log-transformed expression. For human ovarian supporting cells only, 'o' denotes TF whose binding motifs are differentially accessible (that is, TF can bind their potential targets); 'a' denotes TF whose targets are also differentially expressed (that is, differentially activated TF) and asterisk denotes TF that meets both 'o' and 'a' conditions. Conservation heatmap (right) highlights significant overexpression (log2 fold change > 0 and FDR < 0.05) in each species. TFs whose upregulation is conserved across species are highlighted with bold/coloured labels. d, Dot plots showing scaled z scored expression of genes coding for interacting ligand–receptor proteins (CellPhoneDB) in supporting and germ cell states in the outer cortex, inner cortex and primordial follicles. Specific interacting partners are linked with a matching symbol. CoelEpi, coelomic epithelium; Expr, expressed; FGC, fetal germ cells; preGC, pregranulosa cells; granulosa, developing granulosa.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
