Diverse monogenic subforms of human spermatogenic failure
Nagirnaja L, Lopes AM, Charng WL, Miller B, Stakaitis R, Golubickaite I, Stendahl A, Luan T, Friedrich C, Mahyari E, Fadial E, Kasak L, Vigh-Conrad K, Oud MS, Xavier MJ, Cheers SR, James ER, Guo J, Jenkins TG, Riera-Escamilla A, Barros A, Carvalho F, Fernandes S, Gonçalves J, Gurnett CA, Jørgensen N, Jezek D, Jungheim ES, Kliesch S, McLachlan RI, Omurtag KR, Pilatz A, Sandlow JI, Smith J, Eisenberg ML, Hotaling JM, Jarvi KA, Punab M, Rajpert-De Meyts E, Carrell DT, Krausz C, Laan M, O'Bryan MK, Schlegel PN, Tüttelmann F, Veltman JA, Almstrup K, Aston KI, Conrad DF, 26.12.2022
Abstract
Non-obstructive azoospermia (NOA) is the most severe form of male infertility and typically incurable. Defining the genetic basis of NOA has proven challenging, and the most advanced classification of NOA subforms is not based on genetics, but simple description of testis histology. In this study, we exome-sequenced over 1000 clinically diagnosed NOA cases and identified a plausible recessive Mendelian cause in 20%. We find further support for 21 genes in a 2-stage burden test with 2072 cases and 11,587 fertile controls. The disrupted genes are primarily on the autosomes, enriched for undescribed human "knockouts", and, for the most part, have yet to be linked to a Mendelian trait. Integration with single-cell RNA sequencing data shows that azoospermia genes can be grouped into molecular subforms with synchronized expression patterns, and analogs of these subforms exist in mice. This analysis framework identifies groups of genes with known roles in spermatogenesis but also reveals unrecognized subforms, such as a set of genes expressed across mitotic divisions of differentiating spermatogonia. Our findings highlight NOA as an understudied Mendelian disorder and provide a conceptual structure for organizing the complex genetics of male infertility, which may provide a rational basis for disease classification.
Nagirnaja L, Lopes AM, Charng WL, Miller B, Stakaitis R, Golubickaite I, Stendahl A, Luan T, Friedrich C, Mahyari E, Fadial E, Kasak L, Vigh-Conrad K, Oud MS, Xavier MJ, Cheers SR, James ER, Guo J, Jenkins TG, Riera-Escamilla A, Barros A, Carvalho F, Fernandes S, Gonçalves J, Gurnett CA, Jørgensen N, Jezek D, Jungheim ES, Kliesch S, McLachlan RI, Omurtag KR, Pilatz A, Sandlow JI, Smith J, Eisenberg ML, Hotaling JM, Jarvi KA, Punab M, Rajpert-De Meyts E, Carrell DT, Krausz C, Laan M, O'Bryan MK, Schlegel PN, Tüttelmann F, Veltman JA, Almstrup K, Aston KI, Conrad DF. Diverse monogenic subforms of human spermatogenic failure. Nat Commun. 2022 Dec 26;13(1):7953. doi: 10.1038/s41467-022-35661-z. PMID: 36572685; PMCID: PMC9792524.
Publication: https://doi.org/10.1038/s41467-022-35661-z Repository: https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs003103.v1.p1
 Disclaimer
Disclaimer
The publication Diverse monogenic subforms of human spermatogenic failure by Nagirnaja L, Lopes AM, Charng WL, Miller B, Stakaitis R, Golubickaite I, Stendahl A, Luan T, Friedrich C, Mahyari E, Fadial E, Kasak L, Vigh-Conrad K, Oud MS, Xavier MJ, Cheers SR, James ER, Guo J, Jenkins TG, Riera-Escamilla A, Barros A, Carvalho F, Fernandes S, Gonçalves J, Gurnett CA, Jørgensen N, Jezek D, Jungheim ES, Kliesch S, McLachlan RI, Omurtag KR, Pilatz A, Sandlow JI, Smith J, Eisenberg ML, Hotaling JM, Jarvi KA, Punab M, Rajpert-De Meyts E, Carrell DT, Krausz C, Laan M, O'Bryan MK, Schlegel PN, Tüttelmann F, Veltman JA, Almstrup K, Aston KI, Conrad DF is published under an open access license: https://creativecommons.org/licenses/by-nc/4.0/. Permits non-commercial re-use, distribution, and reproduction in any medium, provided the original work is properly cited.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Variant prioritization and burden testing in NOA cases
Exome: Whole Exome Sequencing
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:0011961: Non-obstructive azoospermia | 924 | Absence of any measurable level of sperm in his semen, resulting from a defect in the production of spermatozoa in the testes. This can be differentiated from obstructive azoospermia on the basis of testicular biopsy. |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | adult | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 924 |
Images
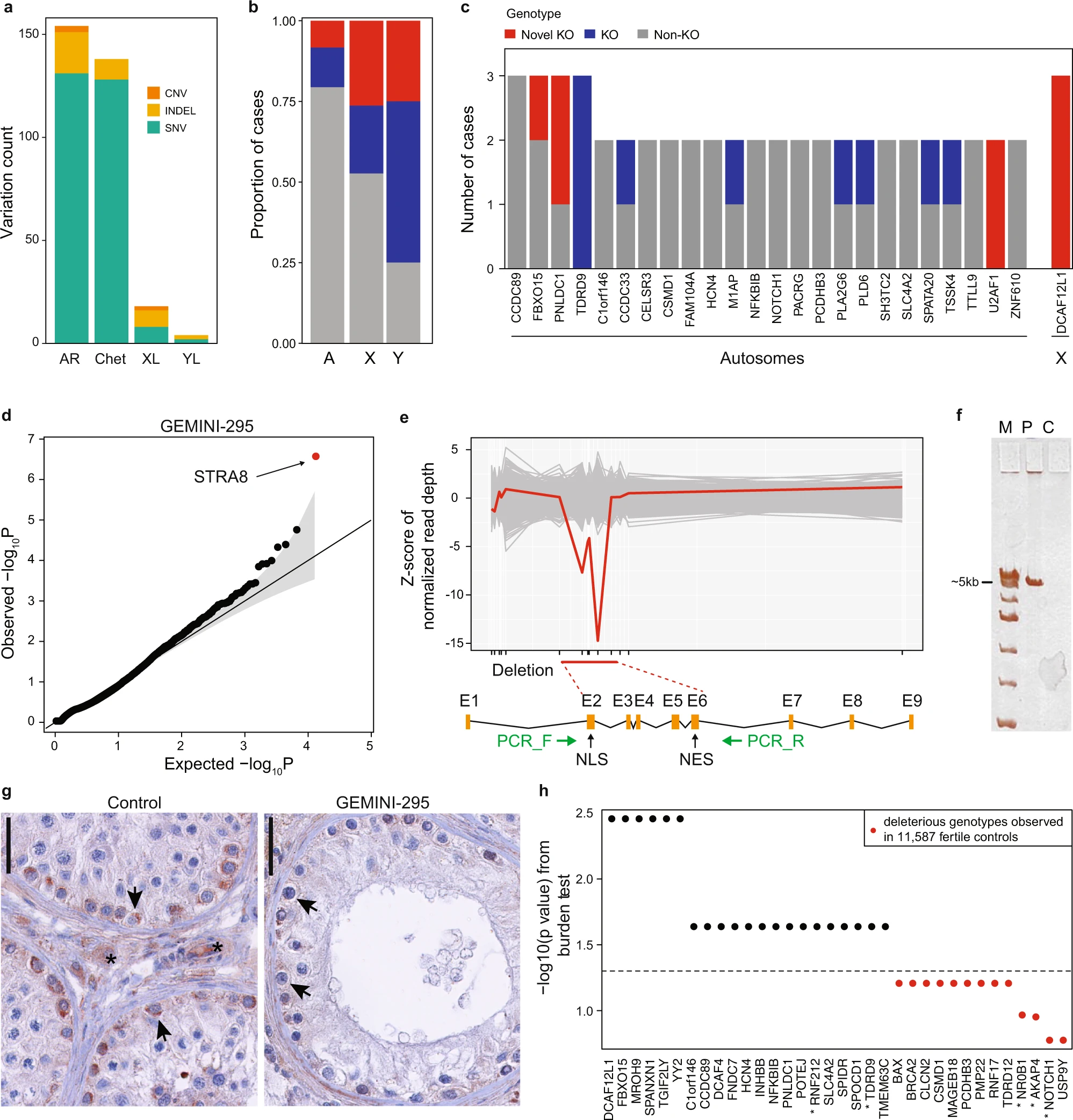
Figure 1: Variant prioritization and burden testing in NOA cases
a Distribution of the prioritized variation types across inheritance modes. AR autosomal recessive, Chet Compound heterozygous, XL X-linked, YL Y-linked. b Predicted hemizygous loss-of-function genotypes appear to be enriched on sex chromosomes compared to biallelic loss-of-function genotypes on autosomes (A) (Fisher’s exact p = 0.018 for chrX and p = 0.032 for chrY vs A). *p < 0.05. c Summary of all genes with multiple case findings in the GEMINI cohort. d PSAP p values of all variants detected in patient GEMINI-295, the carrier of the biallelic STRA8 deletion. STRA8 CNV (the smallest p value) was prioritized as the most likely cause of NOA in GEMINI-295. The gray area represents the 95% confidence interval. e Z-scores of normalized read depth of exome sequencing data around the STRA8 locus, plotted against the STRA8 gene model. NLS nuclear localization; NES, nuclear export signal. Green arrows, PCR primers. f PCR spanning the predicted deletion region yielded a short ~5 kb product in GEMINI-295 (P) validating the homozygous STRA8 deletion. The experiment repeated twice with the same result. C, control; M, ladder. g In control testis, clear staining of STRA8 was observed in spermatogonia (arrows), together with background staining in peritubular and interstitial cells (asterisk). In GEMINI-295, STRA8 protein staining in spermatogonia is much lower (arrows), presumably due to nonsense-mediated decay of the transcript. Three different sections of the controls (n = 8) and the case (n = 1) showed consistent staining patterns. The bar represents 10 µm. h Burden test results from comparison of a combined cohort of 2072 NOA cases with 11,587 fertile controls. Thirty-four genes with prioritized variation in the GEMINI cohort were selected for burden testing; of these 21 were nominally associated with NOA (“Methods”). * indicates genes with at least “limited” clinical validity defined by ref. 7.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 2: Characteristics of NOA genes in the context of male infertility and the broader Mendelian disease landscape
Exome: Whole Exome Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | adult | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 924 |
Images
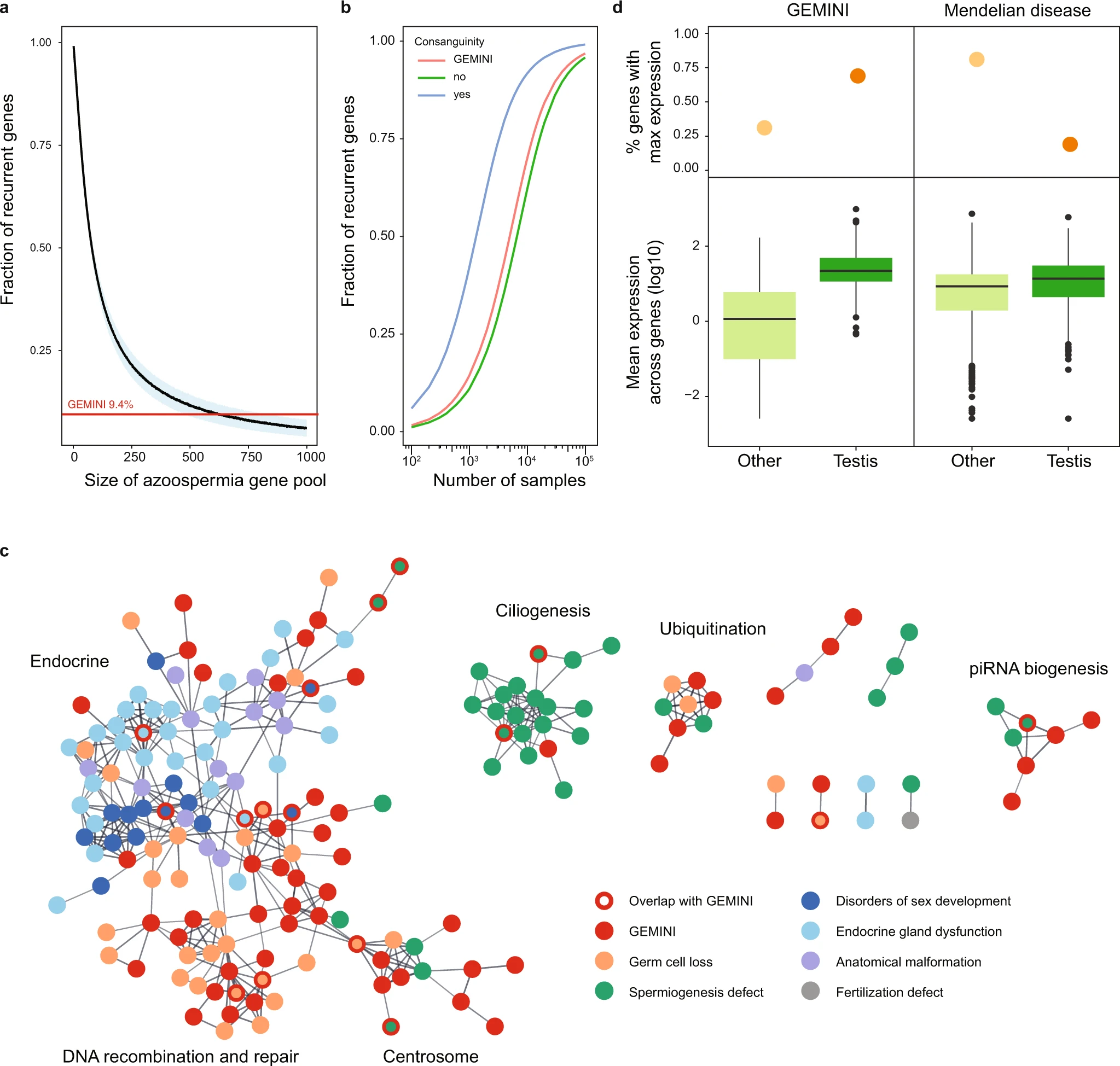
Figure 2: Characteristics of NOA genes in the context of male infertility and the broader Mendelian disease landscape
a Estimation of the total number of azoospermia genes. To detect 9.4% of genes recurrently in non-consanguineous cases with predominantly monogenic NOA, as seen in GEMINI, a gene pool of roughly 625 azoospermia genes would be expected. Blue area represents standard deviation around the mean of 1000 iterations. b Projected number of samples required for observing recurrent gene hits in NOA cohorts considering the estimated pool of 625 azoospermia genes and discovery rates of 76% for consanguineous cases, 20% for GEMINI overall (8% of consanguineous cases) and 15% for non-consanguineous cases. c High confidence protein-protein interactions between GEMINI genes and known male infertility disease genes from Oud et al. 20197 (Supplementary Data 5). d Expression patterns of NOA genes are distinct from that of known recessive Mendelian disease genes (n = 615) in the GTEx atlas of 52 human tissues. NOA genes are more likely to have maximum expression in testis (top panel) and greater testis-specific expression (lower panel). Center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range; points, outliers.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 3: Integrative analysis with testis scRNAseq reveals ‘molecular subforms’ of NOA
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human | 12 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000020: spermatogonium | An euploid male germ cell of an early stage of spermatogenesis. | Human | |||
| CL_0000017: spermatocyte | A male germ cell that develops from spermatogonia. The euploid primary spermatocytes undergo meiosis and give rise to the haploid secondary spermatocytes which in turn give rise to spermatids. | Human | |||
| CL_0000018: spermatid | A male germ cell that develops from the haploid secondary spermatocytes. Without further division, spermatids undergo structural changes and give rise to spermatozoa. | Human | |||
| CL_0000216: Sertoli cell | A supporting cell projecting inward from the basement membrane of seminiferous tubules. They surround and nourish the developing male germ cells and secrete androgen binding protein. Their tight junctions with the spermatogonia and spermatocytes provide a blood-testis barrier. | Human | |||
| CL_0000178: Leydig cell | A Leydig cell is a testosterone-secreting cell in the interstitial area, between the seminiferous tubules, in the testis. | Human | |||
| CL_0000115: endothelial cell | An endothelial cell comprises the outermost layer or lining of anatomical structures and can be squamous or cuboidal. In mammals, endothelial cell has vimentin filaments and is derived from the mesoderm. | Human | |||
| CL_0000235: macrophage | A mononuclear phagocyte present in variety of tissues, typically differentiated from monocytes, capable of phagocytosing a variety of extracellular particulate material, including immune complexes, microorganisms, and dead cells. | Human | |||
| CL_0000236: B cell | A lymphocyte of B lineage that is capable of B cell mediated immunity. | Human | |||
| CL_0000084: T cell | A type of lymphocyte whose defining characteristic is the expression of a T cell receptor complex. | Human | |||
| CL_0000763: myeloid cell | A cell of the monocyte, granulocyte, mast cell, megakaryocyte, or erythroid lineage. | Human |
Images
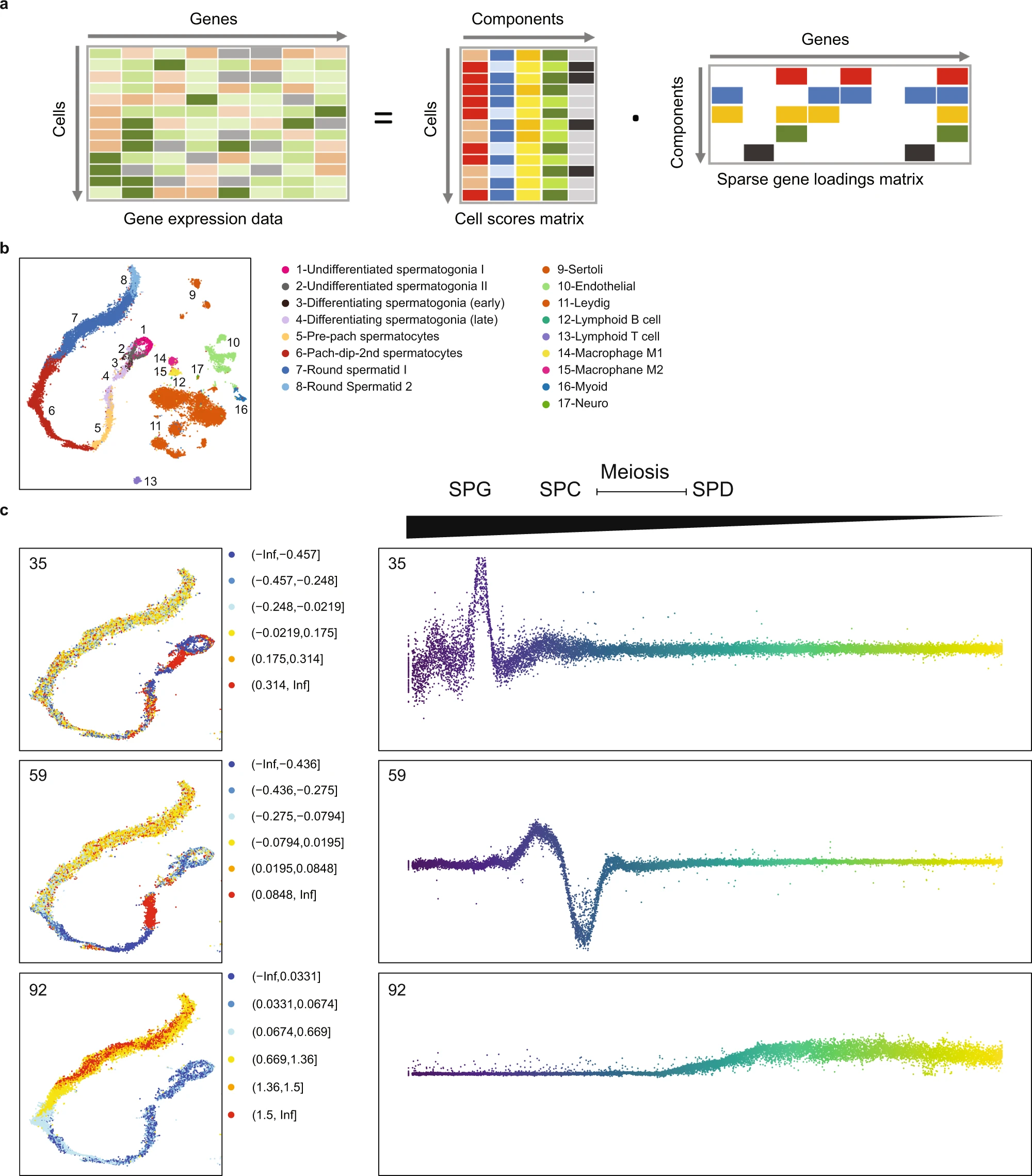
Figure 3: Decomposition of testis gene expression patterns with sparse decomposition of arrays (SDA)
a We applied sparse decomposition analysis (SDA) to testis scRNA-seq data from 12 human donors to identify latent factors (‘components’) representing gene modules. These components are defined by two vectors – one that indicates the loading of each cell on the component, and one that indicates the loading of each gene on the component. b The same scRNA-seq data was summarized using a conventional tSNE analysis, and testicular cell types labels assigned to all cells. c By plotting the cell scores for three representative germ cell components on the tSNE, and as a function of pseudotime, it is apparent that transcription during spermatogenesis can be modeled as series of components overlapping in time, coming on and off gradually on different timescales29. Shown are components with activity that start in spermatogonia (35), spermatocytes (59), and spermatids (92).
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
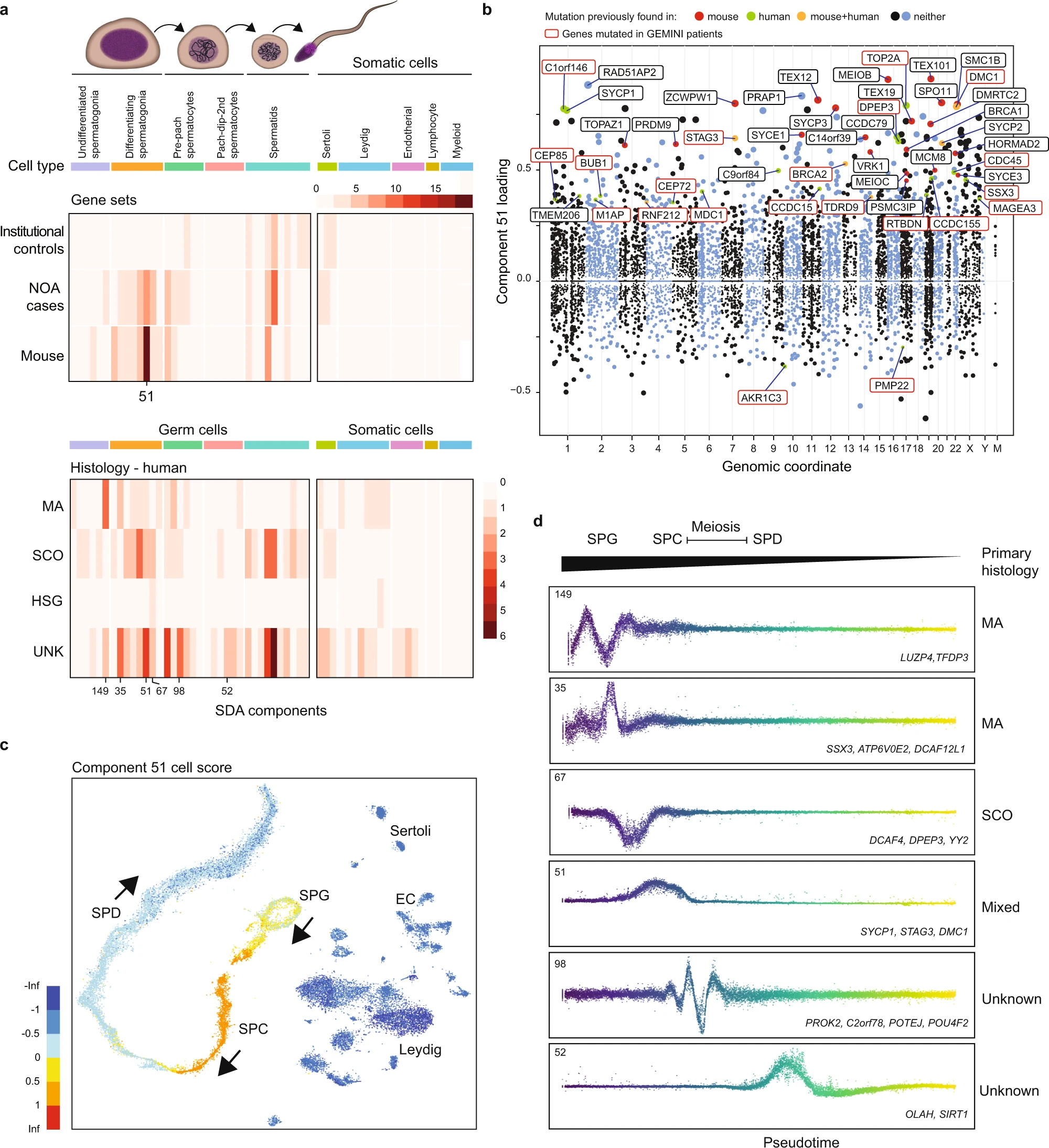
Figure 4: Using SDA components to define molecular subforms of genetic infertility
a Heatmaps summarizing the count of genes loading on each SDA component. Top: The distribution of NOA candidate genes across components is more similar to the distribution of mouse infertility genes, compared to genes with rare damaging genotypes in population controls (p = 3 × 10−4, Pearson and Filon’s Z, two-sided). Bottom: Distribution of NOA candidate genes by testicular histology across SDA components. Genes found in patients with MA show a different pattern of expression compared to genes in SCO patients. SPG spermatogonia, SPC spermatocytes, SPD spermatids, UNK undetermined histology. b Expression of SDA component 51 visualized on a t-SNE representation of the human testis scRNA-seq dataset. This component is expressed in spermatogonia, and more highly in pre-leptotene spermatocytes. c Gene loadings for SDA component 51. Gene loadings reflect which genes are active in a component, with stronger positive or negative loadings indicating greater expression changes in the component compared to the total dataset. d Germ cell expression of components with multiple gene loadings ordered by pseudotime, shown with representative genes and patient histologies.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
Data set 4: Disruption of piRNA biogenesis causes spermatogenic failure in men
Other: Other
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | A typically paired male reproductive gland that produces sperm and that in most mammals is contained within the scrotum at sexual maturity. | Human |
Images

Figure 5: Disruption of piRNA biogenesis causes spermatogenic failure in men
a A schematic of piRNA biogenesis with the components affected among NOA cases in this study highlighted in red. piRNAs are produced by two biogenesis pathways. The primary pathway involves transcription of long precursor-transcripts from genomic clusters, which are then processed in the cytoplasm. b Cell scores for component 59, which encodes both piRNA processing genes and target pre-pre-piRNAs, indicate that it is expressed primarily in pre-leptotene spermatocytes. c Gene loadings for component 59. Note TDRD10 (black box), a protein-coding gene that has yet to be characterized for a role in mammalian piRNA processing. d H&E stain of TDRD9 patient biopsy showing spermatogenic arrest. The most mature germ cell observed were early round spermatids, which often appeared multinucleated (arrows). In addition, many pyknotic cells were observed (arrowheads). Scale bar = 50 µm. e Size distribution of piRNAs detected in the NOA case with a biallelic missense variant in TDRD12 and matching control, derived from small RNA-sequencing of testis tissue. f Significantly decreased fraction of mature (<32 bases) piRNAs was detected in testis of patients with piRNA pathway mutations, indicative of faulty processing of the immature piRNA transcripts.
Licensed under: https://creativecommons.org/licenses/by-nc/4.0/
