Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors
Di Chen, Na Sun, Lei Hou, Rachel Kim, Jared Faith, Marianna Aslanyan, Yu Tao, Yi Zheng, Jianping Fu, Wanlu Li, Manolis Kellis, Amander Clark, 24.12.2019
Abstract
In vitro gametogenesis is the process of making germline cells from human pluripotent stem cells. The foundation of this model is the quality of the first progenitors called primordial germ cells (PGCs), which in vivo are specified during the peri-implantation window of human development. Here, we show that human PGC (hPGC) specification begins at day 12 post-fertilization. Using single-cell RNA sequencing of hPGC-like cells (hPGCLCs) differentiated from pluripotent stem cells, we discovered that hPGCLC specification involves resetting pluripotency toward a transitional state with shared characteristics between naive and primed pluripotency, followed by differentiation into lineage-primed TFAP2A+ progenitors. Applying the germline trajectory to TFAP2C mutants reveals that TFAP2C functions in the TFAP2A+ progenitors upstream of PRDM1 to regulate the expression of SOX17. This serves to protect hPGCLCs from crossing the Weismann’s barrier to adopt somatic cell fates and, therefore, is an essential mechanism for successfully initiating in vitro gametogenesis.
CHEN, Di, et al. Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors. Cell reports, 2019, 29. Jg., Nr. 13, S. 4568-4582. e5.
Publication: https://doi.org/10.1016/j.celrep.2019.11.083 Repository: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140021
 Disclaimer
Disclaimer
The publication Human Primordial Germ Cells Are Specified from Lineage-Primed Progenitors by Di Chen, Na Sun, Lei Hou, Rachel Kim, Jared Faith, Marianna Aslanyan, Yu Tao, Yi Zheng, Jianping Fu, Wanlu Li, Manolis Kellis, Amander Clark is published under an open access no derivatives license: : https://creativecommons.org/licenses/by-nc-nd/4.0/. Granted rights: share — copy and redistribute the material in any medium or format. No alterations allowed. Thus, tables can not be shown in MFGA format.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Ex-vivo: Tracing the Specification of hPGCs
Proteome: Other
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:control | 4-5 week human Embryo used as positive control for identifying hPGCs |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001099: blastocyst | different stages | Human | 127 | |
| BTO_0000379: embryo | Embryonic | day 5 or day 6 post-fertilization embryos used for embryo attachment culture | Human | 78 |
Images
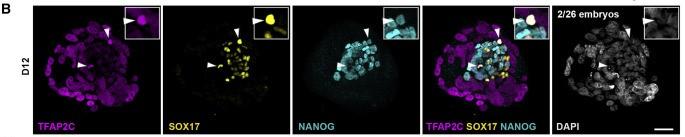
Figure 1 B: Human embryo attachment culture staining
Human embryo attachment culture at day 12 stained for TFAP2C (purple), SOX17 (yellow), and NANOG (cyan). Two TFAP2C/SOX17/NANOG triple-positive cells are highlighted by a white arrowhead, one of which is enlarged in inset. The number in the 4′,6-diamidino-2-phenylindole (DAPI) (gray) panel indicates triple-positive cells were identified in two day 12 embryos from a total of 26 embryos examined at this time point.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
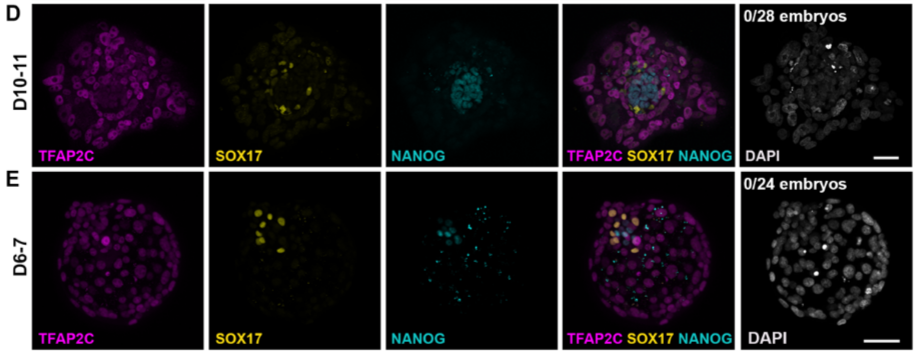
Figure S1 D and E:Human PGCs can be identified by triple staining for NANOG/SOX17/TFAP2C and UMAP display of scRNA-seq cells, Related to Figure 1
No triple positive cells were identified from D6-7 human blastocysts or D10-11 attachment cultured human embryos.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 2: In vitro: Tracing the specification of hPGCs using hPGCL cell culture
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0002322: embryonic stem cell | Embryonic | stem cell line UCLA1 and UCLA2 | Human | ||
| CL_0002371: somatic cell | Aggregate cells at days 1–4 | Human | |||
| incipient mesoderm-like cells (iMeLCs) | Human |
Images
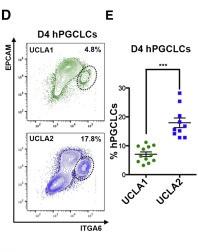
Figure 1 D and E: Identification and Percentages of ITGA6/EPCAM double-positive hPGCLCs in UCLA1 and UCLA2
D: Identification of ITGA6/EPCAM double-positive hPGCLCs by flow cytometry from UCLA1 and UCLA2.</br>E:Percentage of ITGA6/EPCAM double-positive hPGCLCs isolated from UCLA1 and UCLA2 at day 4 of differentiation. ∗∗∗, indicates statistically significant difference comparing more than 10 biological replicates of UCLA1 and UCLA2 at day 4 of differentiation (shown is mean and SEM).
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
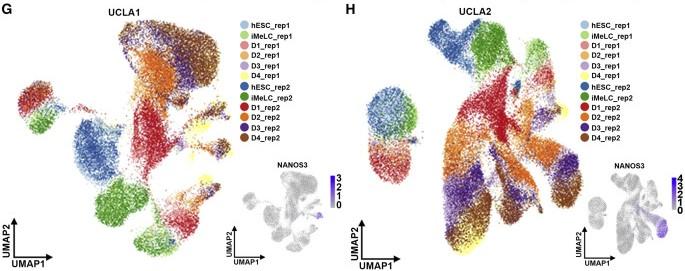
Figure 1 G and H: Tracking the Specification of Human PGCs
UMAP display of all cells in the UCLA1 (G) and UCLA2 (H) dataset in biological duplicate (n = 12 samples for each cell line). The expression of NANOS3 indicates hPGCLCs. Scale bars: 50 μM. See also Figure S1.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
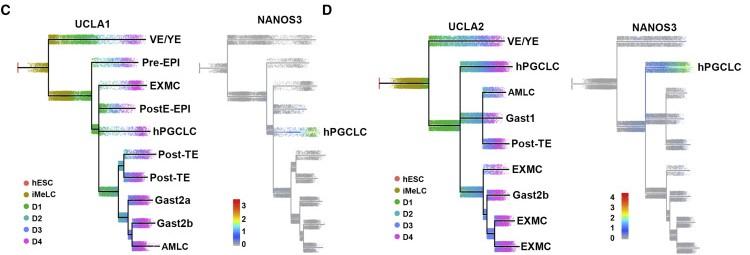
Figure 2 C and D: URD tree showing the developmental trajectories from hESC and iMeLCs through 4 days of aggregate differentiation from UCLA1 (C) and UCLA2 (D)
The expression of NANOS3 marks the end of the germline trajectory and establishment of hPGCLCs in (C) and (D). Cells with endoderm (Endo) identity are outside of the germline trajectory, indicating hPGCLCs do not originate from SOX17-positive endoderm. VE/YE, visceral/yolk-sac endoderm; EXMC, extra-embryonic mesenchyme; Pre-EPIs, pre-implantation epiblasts; PostE-EPI, post-implantation early epiblasts; Post-TE, post-implantation trophectoderm; Gast, gastrulating cells; AMLCs, amnion-like cells (Nakamura et al., 2016, Zheng et al., 2019).See also Figure S2.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 3: hPGCLCs Are Specified through a Pluripotent State with Both Naive and Primed Characteristics
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0002322: embryonic stem cell | Embryonic | hESC UCLA1 | Human | ||
| CL_0002322: embryonic stem cell | Embryonic | hESC UCLA2 | Human |
Images
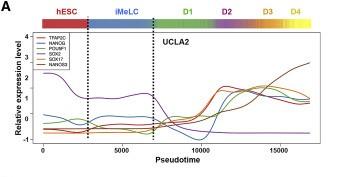
Figure 3 A: Expression patterns during germline tajectory
The expression patterns of TFAP2C, NANOG, SOX2, POU5F1(OCT4), SOX17, and NANOS3 within the germline trajectory of UCLA2. Dash lines outline iMeLCs in germline trajectory in (A)–(C)
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
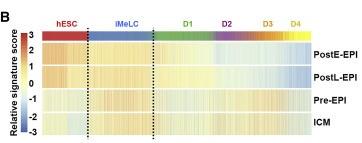
Figure 3 B: Specification through a Translational Germinal Pluripotent State
Relative average expression of signature genes from cynomolgus ICM (inner cell mass), Pre-EPIs, PostE-EPIs, and PostL-EPI (post-implantation late epiblast) (Nakamura et al., 2016) in germline trajectory of UCLA2.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
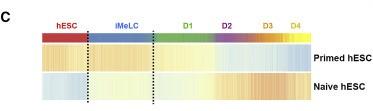
Figure 3 C: iMeLCs in the germline trajectory showed signatures of both naive and primed hESCs
Relative average expression of signature genes from primed and naive hESCs (Messmer et al., 2019) in germline trajectory of UCLA2. Signature score bar is same as (B).
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
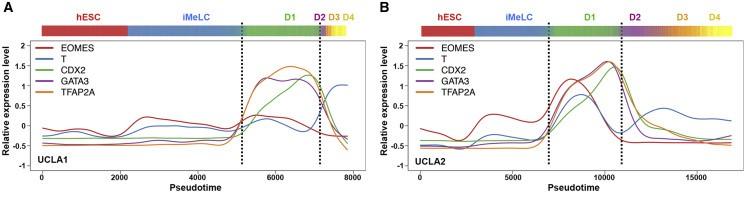
Figure 4 A and B: hPGCLC Specification Involves a Trajectory through a Transient TFAP2A-Positive State with a Signature of Both Amnion and Gastrulating Cell Fates
The expression patterns of EMOES, T, CDX2, GATA3, and TFAP2A within germline trajectory of UCLA1 (A) and UCLA2 (B).
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 4: TFAP2C Functions Upstream of SOX17 and Safeguards Germ Cell Fate in the Germline Trajectory
Transcriptome: Other
Species
| Species |
|---|
| Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0002322: embryonic stem cell | Embryonic | (wildtype) hESC UCLA1 | Human | ||
| CL_0002322: embryonic stem cell | Embryonic | mutant hESC UCLA1 TFAP2C−/− | Human | ||
| CL_0002322: embryonic stem cell | Embryonic | hESC UCLA2 | Human |
Images
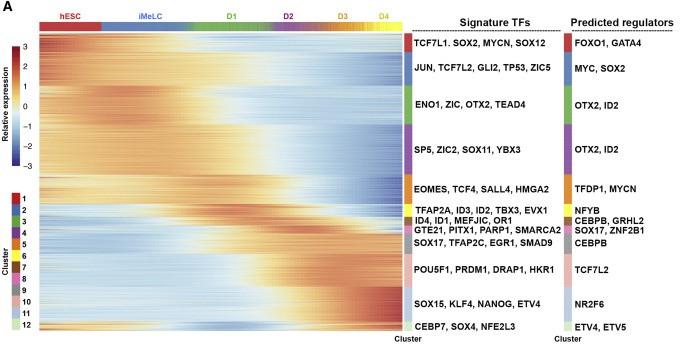
Figure 5 A: . TFAP2C Acts Upstream of SOX17
Heatmap showing the differentially expressed genes in the UCLA2 germline trajectory. Signature transcription factors (TFs) and predicted regulators are shown for each cluster.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
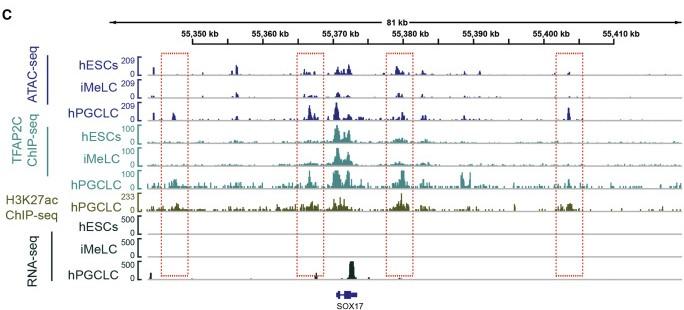
Figure 5 C: TFAP2C Acts Upstream of SOX17 via ATAC-seq, TFAP2C ChIP-seq, H3K27ac ChIP-seq, and RNA-seq
Tracks showing the ATAC-seq, TFAP2C ChIP-seq, H3K27ac ChIP-seq, and RNA-seq peaks around SOX17 site at each stage. See also Figure S5.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
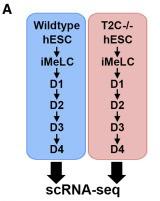
Figure 6 A: Experimental Setup TFAP2C Wildtype/ mutant
Experimental design comparing scRNA-seq libraries of TFAP2C mutant (T2C−/−) hESCs, iMeLCs, day 1, day 2, day 3, and day 4 to wild-type cells of the same genetic background (UCLA1).
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
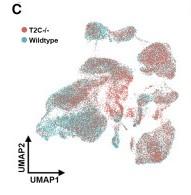
Figure 6 C: Overlay from wild-type UCLA1 (blue) and T2C−/− cells (pink)
Overlay from each of the six time points. Clusters that are largely pink are enriched in mutant cells; clusters that are largely blue are enriched in wild-type cells.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
