Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs
Shino Mitsunaga, Junko Odajima, Shiomi Yawata, Keiko Shioda, Chie Owa, Kurt J. Isselbacher, Jacob H. Hanna, and Toshi Shioda, 30.10.2017
Abstract
Pluripotent stem cell-derived human primordial germ cell-like cells (hPGCLCs) provide important opportunities to study primordial germ cells (PGCs). We robustly produced CD38+ hPGCLCs [∼43% of FACS-sorted embryoid body (EB) cells] from primed-state induced pluripotent stem cells (iPSCs) after a 72-hour transient incubation in the four chemical inhibitors (4i)-naïve reprogramming medium and showed transcriptional consistency of our hPGCLCs with hPGCLCs generated in previous studies using various and distinct protocols. Both CD38+ hPGCLCs and CD38− EB cells significantly expressed PRDM1 and TFAP2C, although PRDM1 mRNA in CD38− cells lacked the 3′-UTR harboring miRNA binding sites regulating mRNA stability. Genes up-regulated in hPGCLCs were enriched for cell migration genes, and their promoters were enriched for the binding motifs of TFAP2 (which was identified in promoters of T, NANOS3, and SOX17) and the RREB-1 cell adhesion regulator. In EBs, hPGCLCs were identified exclusively in the outermost surface monolayer as dispersed cells or cell aggregates with strong and specific expression of POU5F1/OCT4 protein. Time-lapse live cell imaging revealed active migration of hPGCLCs on Matrigel. Whereas all hPGCLCs strongly expressed the CXCR4 chemotaxis receptor, its ligand CXCL12/SDF1 was not significantly expressed in the whole EBs. Exposure of hPGCLCs to CXCL12/SDF1 induced cell migration genes and antiapoptosis genes. Thus, our study shows that transcriptionally consistent hPGCLCs can be readily produced from hiPSCs after transition of their pluripotency from the primed state using various methods and that hPGCLCs resemble the early-stage PGCs randomly migrating in the midline region of human embryos before initiation of the CXCL12/SDF1-guided chemotaxis.
MITSUNAGA, Shino, et al. Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs. Proceedings of the National Academy of Sciences, 2017, 114. Jg., Nr. 46, S. E9913-E9922.
Publication: https://doi.org/10.1073/pnas.1707779114
 Disclaimer
Disclaimer
The publication Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs by Shino Mitsunaga, Junko Odajima, Shiomi Yawata, Keiko Shioda, Chie Owa, Kurt J. Isselbacher, Jacob H. Hanna, and Toshi Shioda is published under an open access no derivatives license: : https://creativecommons.org/licenses/by-nc-nd/4.0/. Granted rights: share — copy and redistribute the material in any medium or format. No alterations allowed. Thus, tables can not be shown in MFGA format.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Transcriptomal Profiling of CD38+ hPGCLCs Produced via 72-h Pre-EB 4i Reprogramming
Transcriptome: Bulk RNA-Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000379: embryo | Embryonic | Human | ||
| BTO_0002245: foreskin fibroblast cell line | Neonatal | dermal foreskin fibroblast | Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000670: primordial germ cell | Primordial | Human | |||
| Primordial | Human primordial germ cell-like cells (hpGCLCs) | Human | |||
| CL_0002248: pluripotent stem cell | Embryonic | Induced pluripotent stem cells | human | ||
| CL_0002321: embryonic cell | Embryonic | Embryoid body | Human |
Images
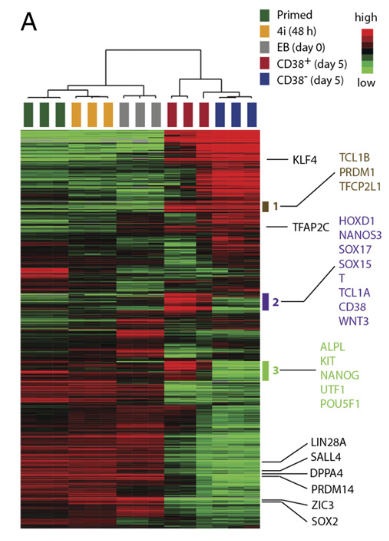
Figure 2 A: Transcriptomal profiles of CD38+ hPGCLCs and precursors
Unsupervised hierarchical clustering of CD38+hPGCLCs and their precursors based on transcriptomes. RNA-seq data were obtained from three independently performed experiments. Gray color indicates “zero value” data, reflecting the absence of expression.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
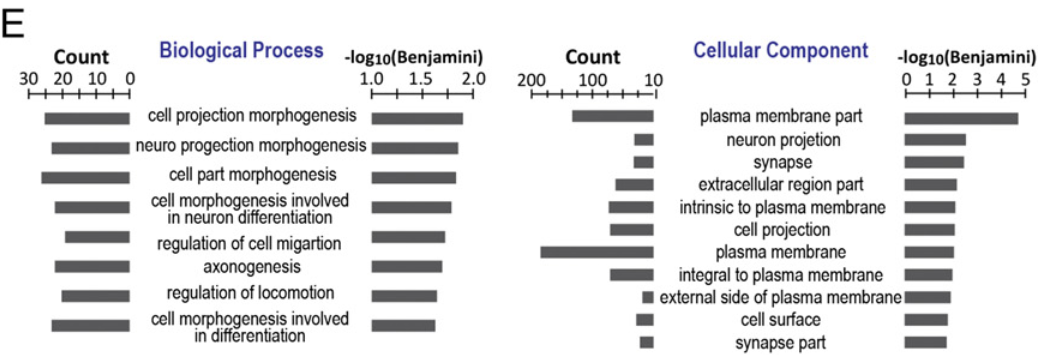
Figure 2 E: Transcriptomal profiles of CD38+ hPGCLCs and precursors
Gene Ontology (GO) analysis of the differentially expressed genes (DEGs) between CD38+ and CD38− EB cells [1,445 genes;false discovery rate (FDR)<5%; more than fourfold change] revealed very strong enrichment of genes involved in cell migration (Fig. 2E,Left) and plasma membrane activities (Fig. 2E,Right), both of which included formation of cell projections.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
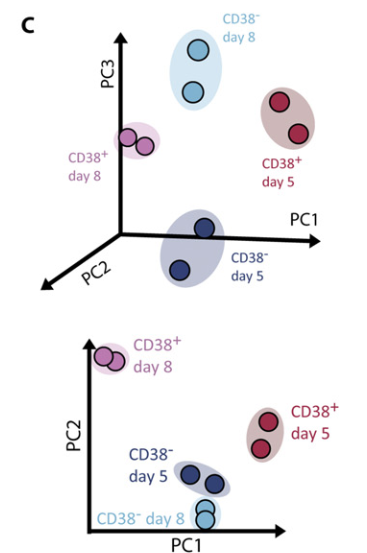
Figure 3 C: Transcriptomal profiles of CD38+ hPGCLCs and precursors
Transcriptomal PCA of 3,500 DEGs with the lowest FDRs revealed a clear distinction between days 5 and 8CD38+hPGCLCs
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 2: Transcriptomal Consistency of hPGCLCs Produced Using Various Protocols and FACS Markers
Transcriptome: Bulk RNA-Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000379: embryo | Embryonic | Human | ||
| BTO_0002245: foreskin fibroblast cell line | Neonatal | Dermal foreskin fibroblasts | Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000670: primordial germ cell | Primordial | Human | |||
| Primordial | Human primordial germ cell like cells (hPGCLCs) | Human | |||
| CL_0002248: pluripotent stem cell | Embryonic | Induced pluripotent stem cells | Human | ||
| CL_0002321: embryonic cell | Embryonic | Embryoid body | Human |
Images
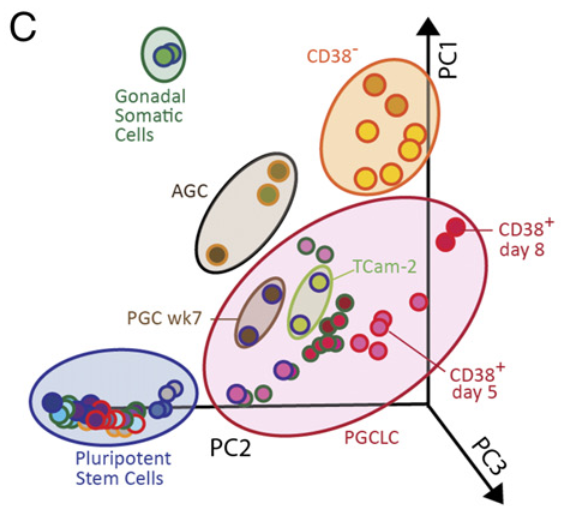
Figure 3 C:Marker gene expression profiles of human germ cells, hPGCLCs, and precursors across various protocols
PCA of human embryonic and in vitro germline cells with data shown in A. PC, principal component.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
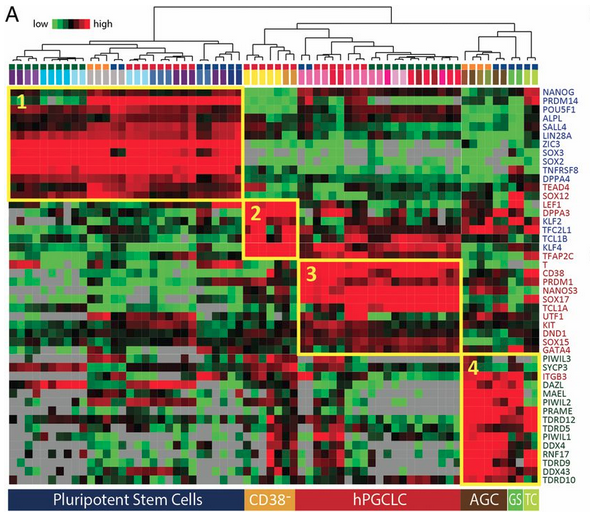
Figure 3 A: Marker gene expression profiles of human germ cells, hPGCLCs, and precursors across various protocols.
Unsupervised hierarchical clustering of human embryonic and in vitro germline cells generated in four independent laboratories using different protocols. Clustering was performed based on 46 marker genes of pluripotency (blue), PGC (red), and AGCs (green). Color-coded cell types shown at the top of the heat map are indicated in B. Groups of genes expressed in the (1) pluripotent precursor cells, (2) nongermline EB cells (CD38−), (3) hPGCLCs, and (4) AGCs are shown with yellow rectangles. Gray color indicates zero value data, reflecting the absence of expression. GS, gonadal somatic cells; TC, TCam-2 human seminoma cells.</br>MFGA curated description: Color identifications</br>This study: red caps</br>Sasaki K, et al. (2015) Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 17:178–194.: green caps</br>Irie N, et al. (2015) SOX17 is a critical specifier of human primordial germ cell fate. Cell 160:253–268: blue caps</br>Gkountela S, et al. (2013) The ontogeny of cKIT+ human primordial germ cells proves to be a resource for human germ line reprogramming, imprint erasure and in vitro differentiation. Nat Cell Biol 15:113–122:orange caps.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
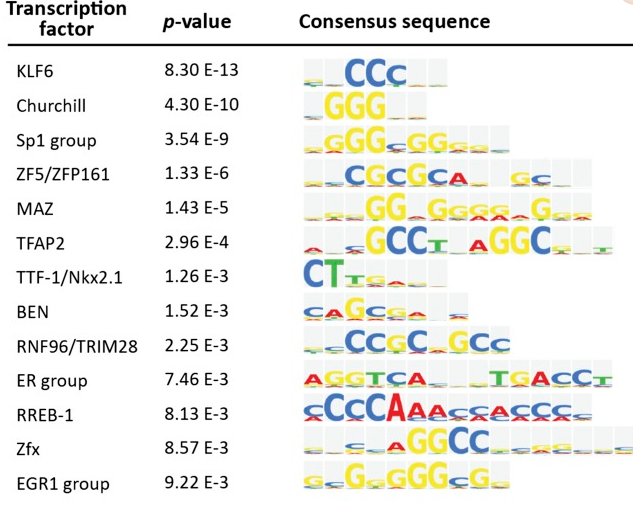
Figure 4 A: List of significantly enriched TFs (P<0.01)
TF binding motifs enriched in the regulatory sequences of 537 DEGs expressed more strongly in CD38+ hPGCLCs than in CD38− EB cells. The P value was defined by the F-match algorithm and calculated by the TRANSFAC server.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
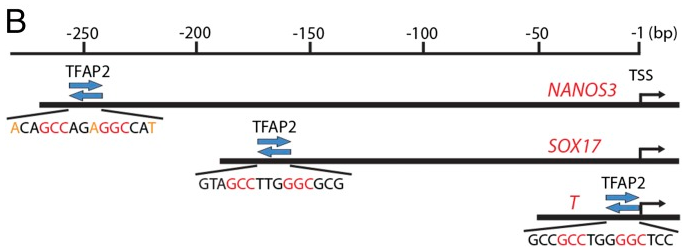
Figure 4 B: Locations of the TFAP2 binding motifs in promoters of NANOS3, SOX17, and T/BRACHYURY
Matches to the core motif (GCCNNNGGC) are shown in red, and matches to the additional contributing sequences are shown in orange.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
Data set 3: Expression of the CXCR4 Chemotactic Cytokine Receptor in CD38+ PGCLCs and Transcriptional Effects of CXCL12
Transcriptome: Bulk RNA-Sequencing
Species
| Species |
|---|
| Human |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0000379: embryo | Embryonic | Human | ||
| BTO_0002245: foreskin fibroblast cell line | Neonatal | Dermal foreskin fibroblasts | Human |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000670: primordial germ cell | Primordial | Human | |||
| Primordial | Human primordial germ cell like cells (hPGCLC) | Human | |||
| CL_0002248: pluripotent stem cell | Embryonic | Induced pluripotent stem cells | Human | ||
| CL_0002321: embryonic cell | Embryonic | Embryoid body | Human |
Images
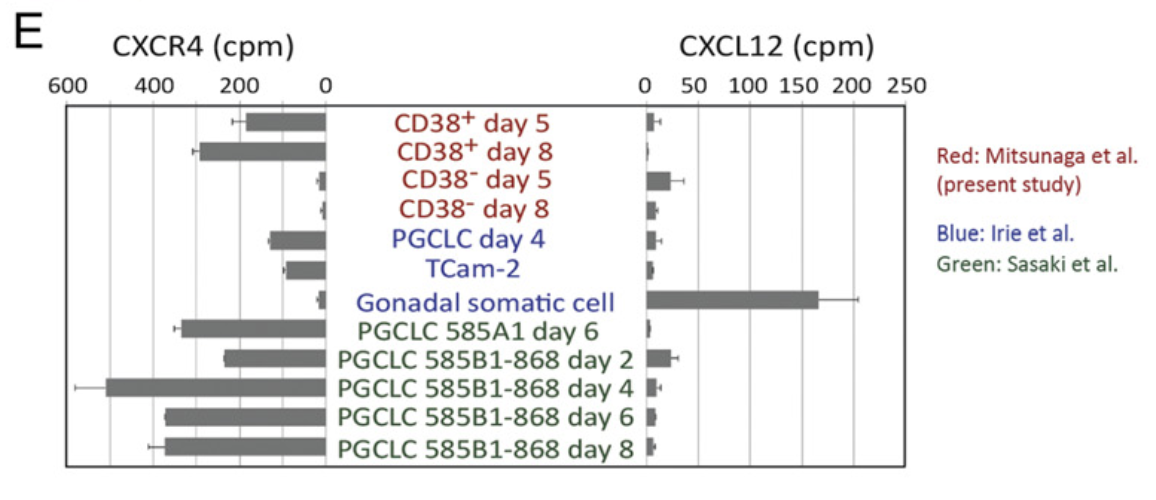
Figure 5 E: Localization of CD38+hPGCLCs at the outermost surface layer of EBs
Expression of the CXCR4 chemotaxis receptor and its ligand CXCL12 in human germline cells and gonadal somatic cells. RNA-seq data generated in four independent laboratories were conormalized to obtain relative expression profiles (of CXCR4 and CXCL12 mRNA across cell types. cpm, Counts per million.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
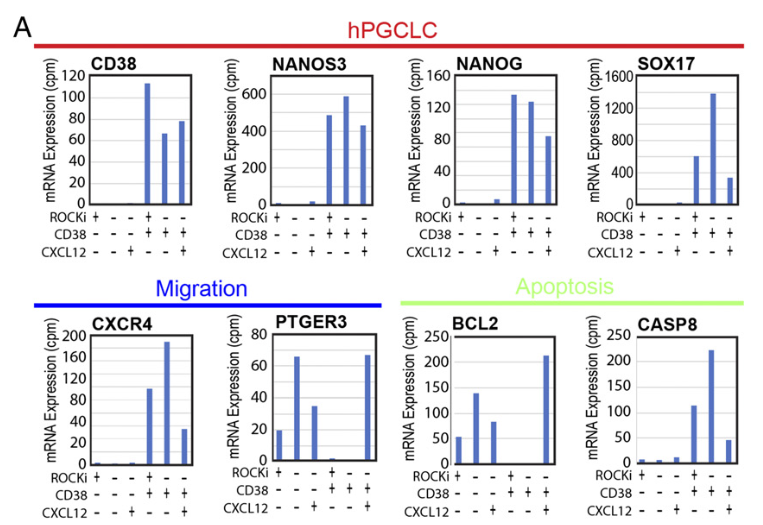
Figure 6 A: Effects of CXCL12 on mRNA expression in hPGCLCs.
Expression of mRNA transcripts in CD38+ hPGCLCs or CD38− EB cells at day 8 EBs. EBs were cultured in the standard condition until day 5 and then incubated for an additional 3 d in the presence or absence of Rho Kinase Inhibitor (ROCKi) and/or CXCL12 in the PGCLC production medium. Amounts of mRNA expression are shown as counts per million (cpm) values of normalized reads of three repeated RNA-seq experiments.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
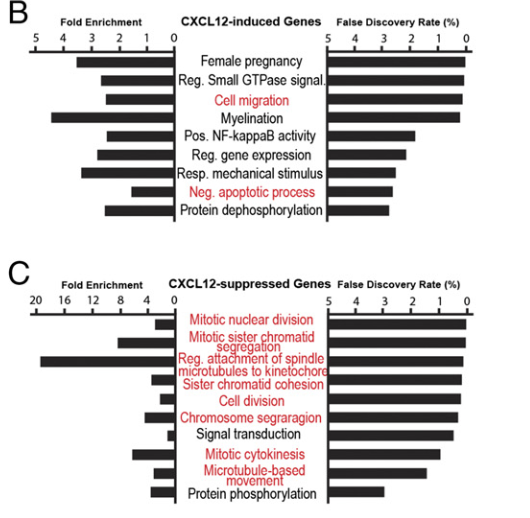
Figure 6 B and C: Effects of CXCL12 on mRNA expression in hPGCLCs.
GO analysis of DEGs in hPGCLCs produced in the ROCKi-deficient medium in the presence or absence of CXCL12. Three RNA-seq experiments determined 1,190 up-regulated (B) and 735 down-regulated (C) DEGs. Statistically significant GO terms (FDR < 5%) are shown.
Licensed under: https://creativecommons.org/licenses/by-nc-nd/4.0/
