A germ cell‐specific ageing pattern in otherwise healthy men
Sandra Laurentino, Jann‐Frederik Cremers, Bernhard Horsthemke, Frank Tüttelmann, Karen Czeloth, Michael Zitzmann, Eva Pohl, Sven Rahmann, Christopher Schröder, Sven Berres, Klaus Redmann, Claudia Krallmann, Stefan Schlatt, Sabine Kliesch, Jörg Gromoll, 20.09.2020
Abstract
Life‐long sperm production leads to the assumption that male fecundity remains unchanged throughout life. However, recently it was shown that paternal age has profound consequences for male fertility and offspring health. Paternal age effects are caused by an accumulation of germ cell mutations over time, causing severe congenital diseases. Apart from these well‐described cases, molecular patterns of ageing in germ cells and their impact on DNA integrity have not been studied in detail. In this study, we aimed to assess the effects of ‘pure’ ageing on male reproductive health and germ cell quality. We assembled a cohort of 198 healthy men (18–84 years) for which end points such as semen and hormone profiles, sexual health and well‐being, and sperm DNA parameters were evaluated. Sperm production and hormonal profiles were maintained at physiological levels over a period of six decades. In contrast, we identified a germ cell‐specific ageing pattern characterized by a steady increase of telomere length in sperm and a sharp increase in sperm DNA instability, particularly after the sixth decade. Importantly, we found sperm DNA methylation changes in 236 regions, mostly nearby genes associated with neuronal development. By in silico analysis, we found that 10 of these regions are located in loci which can potentially escape the first wave of genome‐wide demethylation after fertilization. In conclusion, human male germ cells present a unique germline‐specific ageing process, which likely results in diminished fecundity in elderly men and poorer health prognosis for their offspring.
LAURENTINO, Sandra, et al. A germ cell‐specific ageing pattern in otherwise healthy men. Aging cell, 2020, 19. Jg., Nr. 10, S. e13242.
Publication: https://doi.org/10.1111/acel.13242
 Disclaimer
Disclaimer
The publication A germ cell‐specific ageing pattern in otherwise healthy men by Sandra Laurentino, Jann‐Frederik Cremers, Bernhard Horsthemke, Frank Tüttelmann, Karen Czeloth, Michael Zitzmann, Eva Pohl, Sven Rahmann, Christopher Schröder, Sven Berres, Klaus Redmann, Claudia Krallmann, Stefan Schlatt, Sabine Kliesch, Jörg Gromoll is published under an open access license: https://creativecommons.org/licenses/by/4.0/. Share — copy and redistribute the material in any medium or format. Adapt — remix, transform, and build upon the material for any purpose, even commercially.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Sperm DNA shows widespread DNA methylation changes with age
Methylome: Whole Genome Bisulfite Sequencing
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:control | 12 | Whole genome bisulphite sequencing on sperm DNA was performed on six of the youngest and the six oldest probands in our study (18–25 years vs. older than 65 years). |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm | Human |
Images
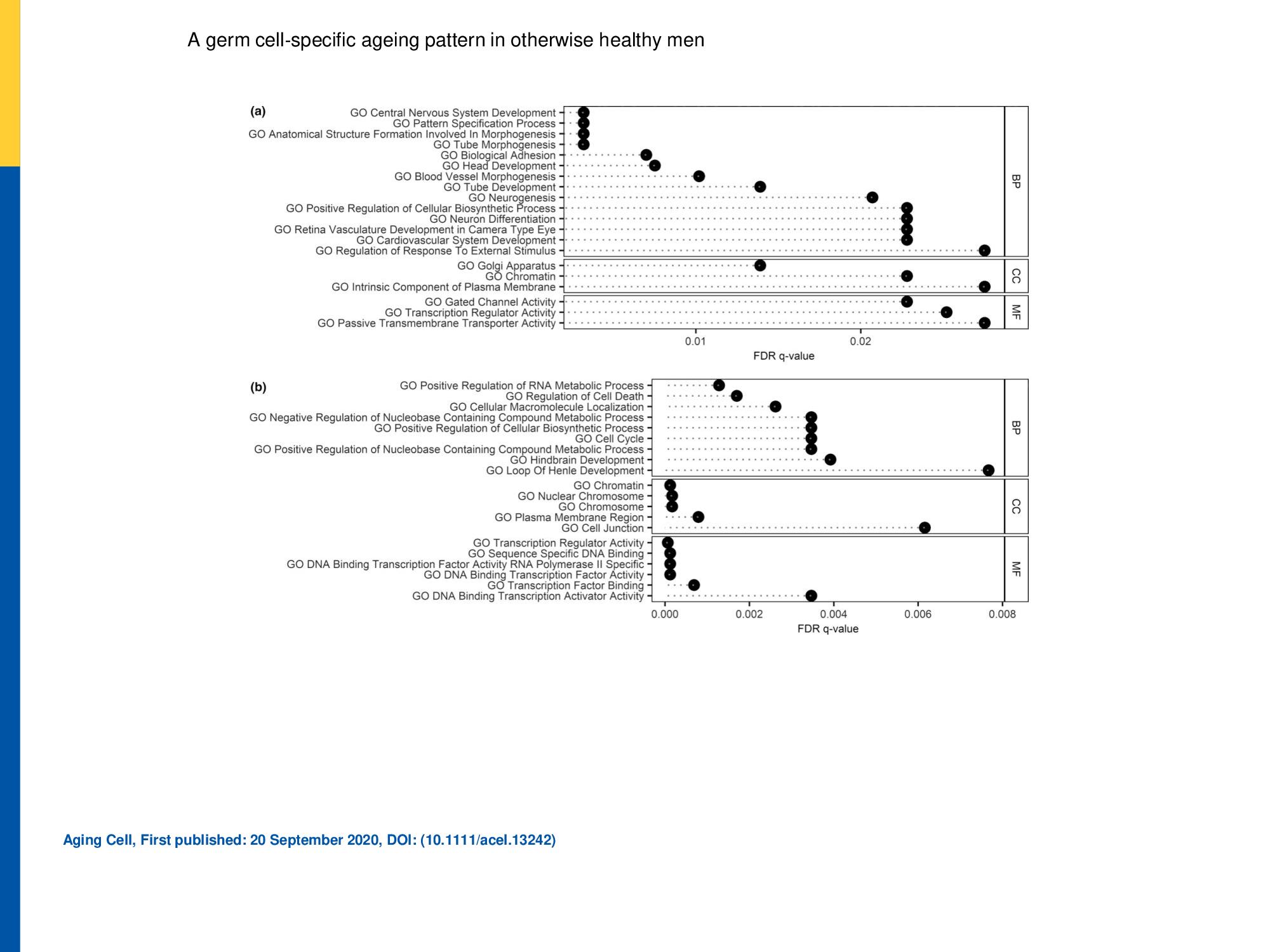
Biological impact of identified hyper‐ and hypomethylated DMRs.
Biological impact of identified hyper‐ (a) and hypomethylated (b) DMRs. Overlap of the DMR‐associated gene list with gene ontology (GO) data sets is shown along with the false discovery rate (FDR) q‐value. The 20 pathways with lowest FDR in the GO domains biological process (BP), cellular component (CC) and molecular function (MF) are displayed
Licensed under: https://creativecommons.org/licenses/by/4.0/
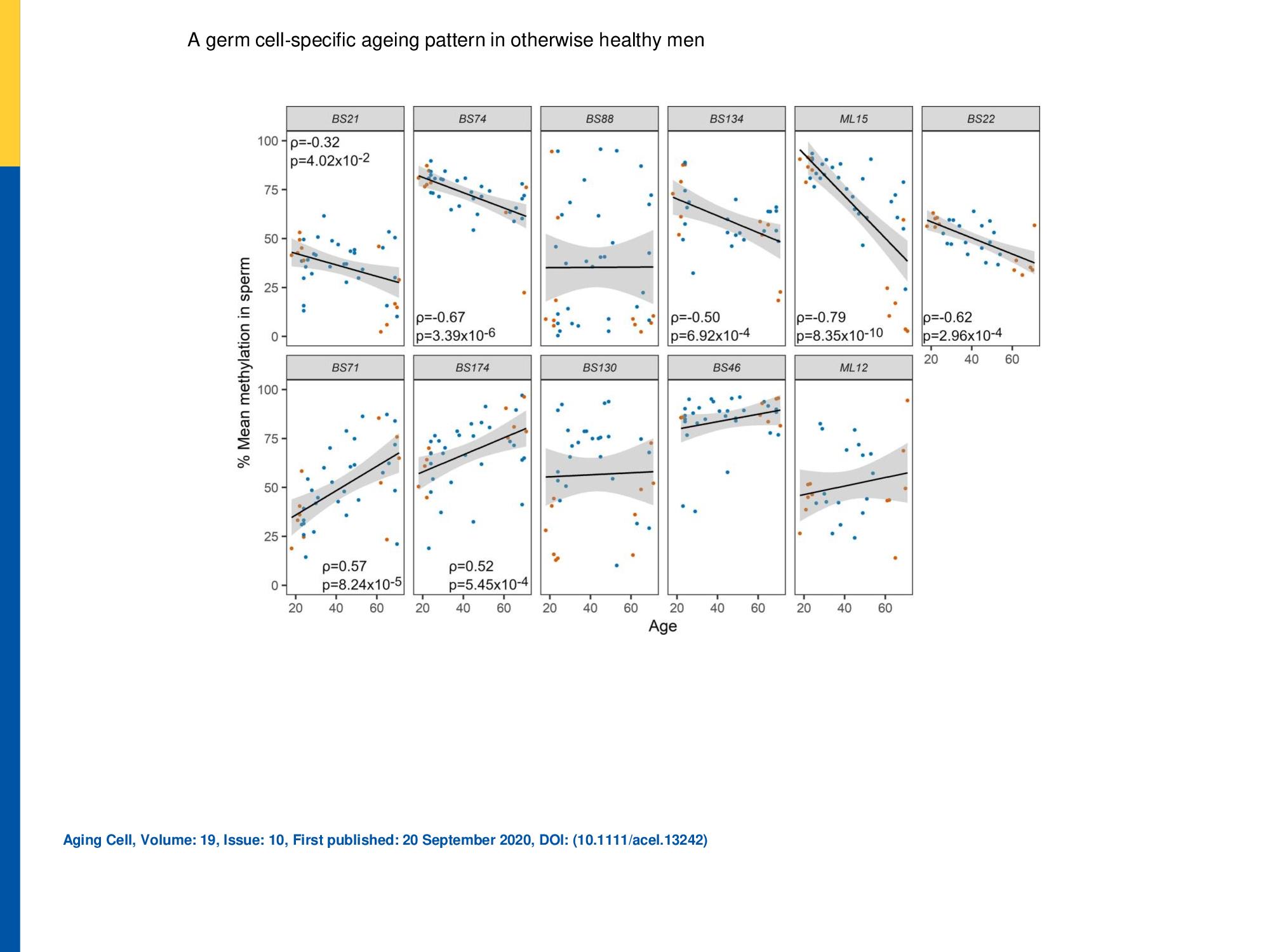
Evaluation of DMRs for age‐dependent changes by DBS.
Eleven age‐associated differentially methylated regions (DMRs) identified by WGBS of sperm of young (G1) and old (G5‐6) men were further analysed in all age groups by DBS in a validation cohort (n = 42). The upper and lower panels show DMRs showing hypo‐ and hypermethylation with age, respectively. The linear trends and 95% confidence intervals are shown by solid lines and grey shading, respectively. The 12 samples previously pooled for WGBS analysis are shown in orange. A statistically significant correlation (Spearmans rank correlation) with age was detected for seven of these DMRs. Only statistically significant correlation results are displayed in each graph
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 2: Sperm telomere length increases with age
Genome: Other
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:other: Other | 198 | participants of 18–84 years of age. A germ cell-specific ageing pattern was characterized, by a steady increase of telomere length in sperm, and a sharp increase in sperm DNA instability, particularly after the sixth decade |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm |
Images
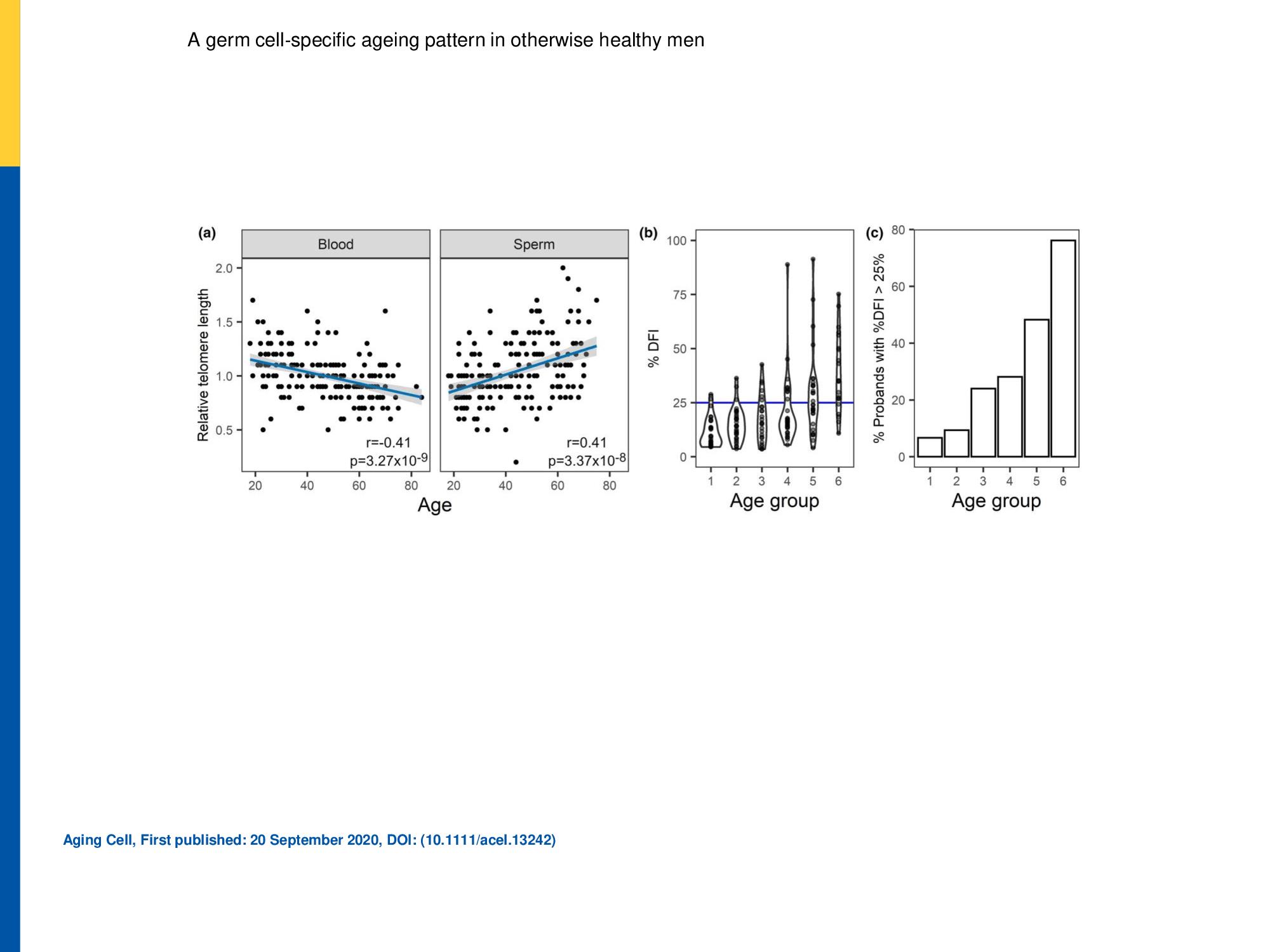
Figure 1. (a) Relative telomere length (b and c) Sperm DNA fragmentation index
(a)Relative telomere length was determined in peripheral blood (n = 194) and swim‐up (n = 179) sperm DNA. Linear trends are displayed in blue and 95% confidence intervals in grey shading. The Pearson correlation coefficients and p‐value are indicated for each case. (b and c) Sperm DNA fragmentation index increases with age. (a) Distribution of DFI values across the age groups is shown in the form of a violin plot. The normal upper value of 25% is marked in blue. (b) The percentage of men in each age group with pathological levels of DFI increases with age
Licensed under: https://creativecommons.org/licenses/by/4.0/
Data set 3: Reproductive parameters in healthy ageing men
Other: Other
Species
| Species |
|---|
| Human |
Conditions
| Human phenotype ontology | Participants | Comment |
|---|---|---|
| HP:other: Other | 197 | Reproductive parameters were evaluated in 197 healthy men |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000019: sperm |
