Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing
S. Lukassen, E. Bosch , A. B. Ekici & A. Winterpacht, 25.04.2018
Abstract
Spermatogenesis in the mouse has been extensively studied for decades. Previous methods, such as histological staining or bulk transcriptome analysis, either lacked resolution at the single-cell level or were focused on a very narrowly defined set of factors. Here, we present the first comprehensive, unbiased single-cell transcriptomic view of mouse spermatogenesis. Our single-cell RNA-seq (scRNAseq) data on over 2,500 cells from the mouse testis improves upon stage marker detection and validation, capturing the continuity of differentiation rather than artificially chosen stages. scRNAseq also enables the analysis of rare cell populations masked in bulk sequencing data and reveals new insights into the regulation of sex chromosomes during spermatogenesis. Our data provide the basis for further studies in the field, for the first time providing a high-resolution reference of transcriptional processes during mouse spermatogenesis.
LUKASSEN, Soeren, et al. Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing. Scientific reports, 2018, 8. Jg., Nr. 1, S. 6521-6521.
Publication: https://doi.org/10.1038/s41598-018-24725-0 Repository: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104556
 Disclaimer
Disclaimer
The publication Characterization of germ cell differentiation in the male mouse through single-cell RNA sequencing by S. Lukassen, E. Bosch , A. B. Ekici & A. Winterpacht is published under an open access license: https://creativecommons.org/licenses/by/4.0/. Permits to share, copy and redistribute the material in any medium or format and to adapt, remix, transform, and build upon the material for any purpose, even commercially.
Curation by the MFGA team Relevant data sets presented in the publication have been identified. If possible, annotations (title, general information, conditions, processed tissue types and processed cell types) have been added based on information from the publication. Data tables and images that provide a good overview on the publication's findings on the data set have been extracted from the publication and/or supplement. If not stated otherwise, images are depicted with title and description exactly as in the publication. Tables have been adjusted to the MFGA table format. Conducted adjustments are explained in the detailed view of the tables. However, titles and descriptions have been adopted from the publication.
Data set 1: Use of scRNA-seq to establish expression profiles of 2,550 germ cells from the adult mouse testis
Transcriptome: Single-cell RNA-Sequencing
Species
| Species |
|---|
| Mouse |
Tissue Types
| BRENDA tissue ontology | Maturity | Description | Species | Replicates |
|---|---|---|---|---|
| BTO_0001363: testis | Adult | 8-week-old C57BL/6J mouse testes | Mouse | 2 |
Cell Types
| Cell ontology | Maturity | Description | Species | Replicates | Cells per replicate |
|---|---|---|---|---|---|
| CL_0000216: Sertoli cell | Adult | Mouse | 2 | ||
| CL_0000178: Leydig cell | Adult | Mouse | 2 | ||
| CL_0000020: spermatogonium | Adult | Spg | Mouse | 2 | |
| CL_0000017: spermatocyte | Adult | SC | Mouse | 2 | |
| CL_0000018: spermatid | Adult | Round Spermatids (RS) | Mouse | 2 | |
| CL_0000018: spermatid | Adult | Elongating Spermatids (ES) | Mouse | 2 | |
| CL_0000018: spermatid | Adult | Condensing Spermatids (CS) | Mouse | 2 |
Images
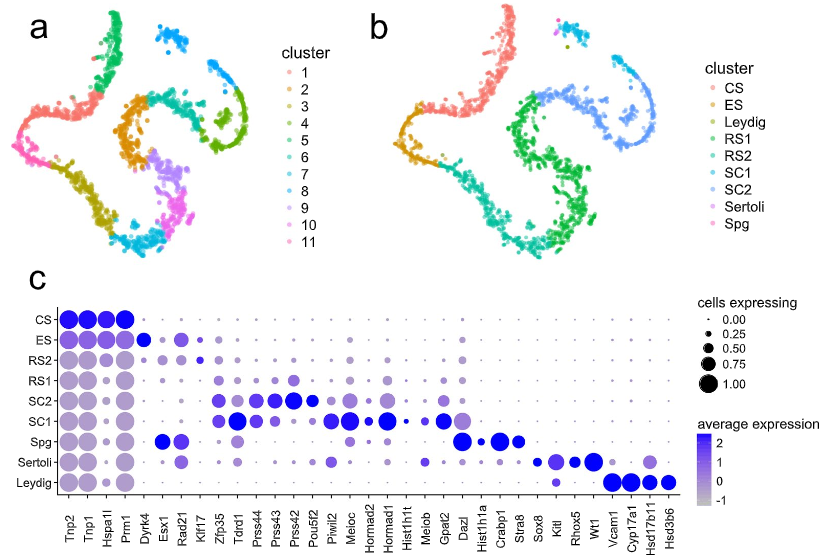
Figure 1: T-SNE projection of cells from both replicates and dot plot of cell expression
(a and b) T-SNE projection of cells from both replicates, colored by cluster identity obtained from (a) graph-based and (b) K-means clustering with K =9. Cell types were assigned through the expression of representative marker genes (Supplementary data Table S3, subfigure c) (c). Dot plot of proportion of cells in the respective K-means cluster expressing each marker (dot size), and average expression (color scale). Spg =spermatogonia, SC = spermatocytes, RS =round spermatids, ES = elongating spermatids, CS =condensing spermatids.
Licensed under: https://creativecommons.org/licenses/by/4.0/
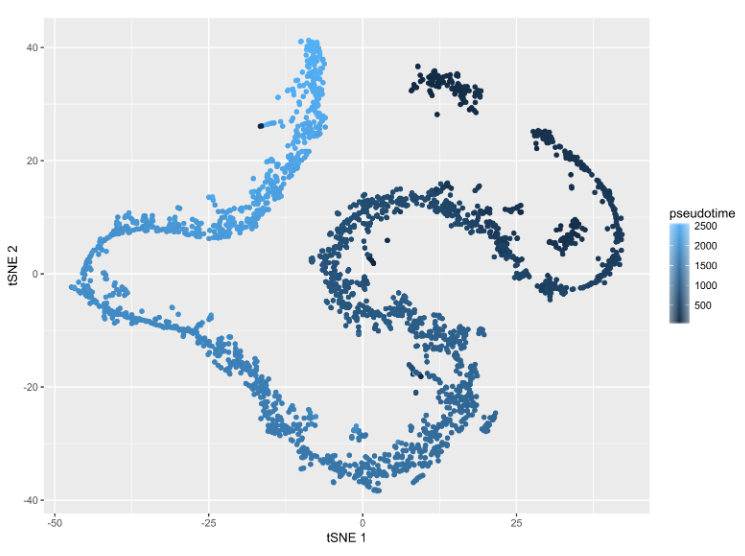
Figure S5: Mapping of the pseudotimerank to the t-SNE projection
Lighter colors indicate later pseudotime.
Licensed under: https://creativecommons.org/licenses/by/4.0/
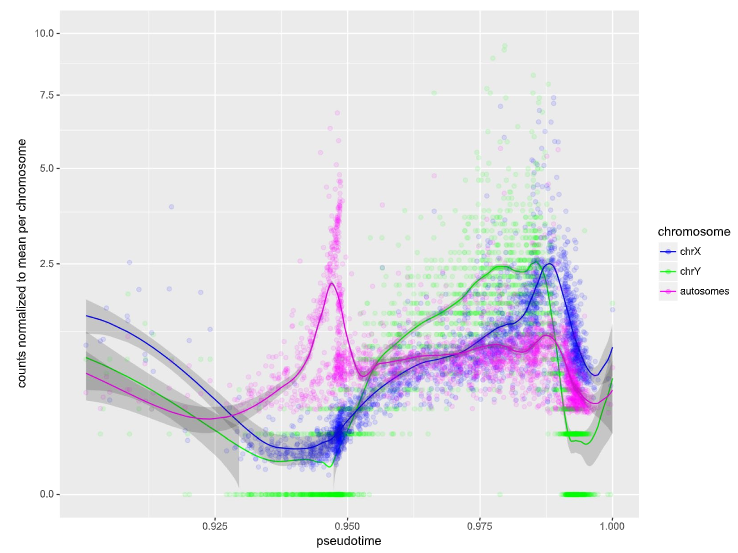
Figure 3: Read counts for autosomes and sex chromosomes, centered on the mean for the respective chromosome.
Individual cells are represented as circles, with smoothed local regression (Loess) represented as solid lines. Shaded areas indicate the standard error of the regression fit. Meiosis occurs roughly between the 0.925 and the 0.95 mark, while Prm1 transcription and thus histone-to-protamine exchange is initiated at approximately 0.986. The fastest decay of the respective transcripts could be observed at 0.991 (Y chromosome) and 0.992 (X chromosome and autosomes).
Licensed under: https://creativecommons.org/licenses/by/4.0/
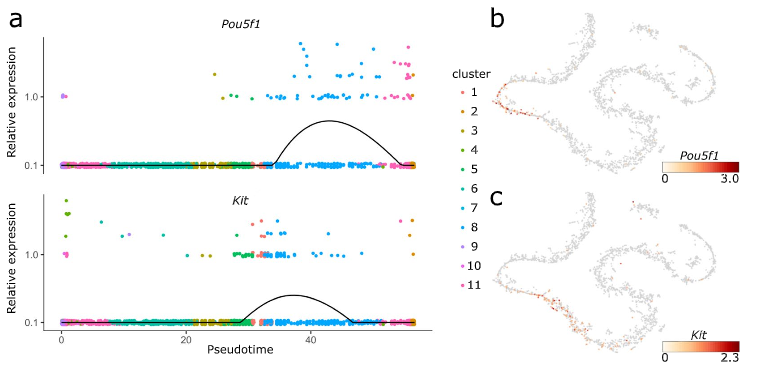
Figure 4: Pou5f and Kit expression
(a) Expression values of Pou5f and Kit along the pseudotime axis. The black line denotes the smoothed, average expression. (b and c) t-SNE projections with colors indicating the expression of Pou5f1 (b) and Kit (c). Our data show that Kit is also expressed at a later stage of spermatogenesis, probably in round spermatids (Fig. 4b).
Licensed under: https://creativecommons.org/licenses/by/4.0/
